1. INTRODUCTION
Knowledge Extraction (KE) is a relatively new research area at the intersection of Data Mining (DM), Information Extraction (IE), and Information Retrieval (IR). The goal of knowledge extraction is to discover knowledge in natural language texts. In terms of knowledge extraction, a variety of types of knowledge can be pulled out from textual data, such as linguistic knowledge and domain-specific lexical and semantic information hidden in unstructured text corpora (Poon & Vanderwende, 2010; Zhou & Zhang, 2007). In light of extracting entities, IE is pertinent to knowledge extraction. It locates specific pieces of data from corpora of natural language texts and populates a relational table from the identified facts. Since the start of the Message Understanding Conferences (MUCs), IE has addressed the issue of transforming unstructured data into structured, relational databases. The transformed text corpus can be mined by various IE techniques such as the application of statistical and machine-learning methods to discover novel relationships in large relational databases.
Developing an IE system is a challenging task. Recently, there has been significant progress in applying data mining methods to help build IE systems (Blaschke, Hirschman, Shatkay, & Valencia, 2010). The major task of these IE systems is entity or relation extraction, such as gene extraction and protein-protein interaction extraction (Airola, Pyysalo, Björne, Pahikkala, Ginter, & Salakoski, 2008; Kim, 2008; Miyao, Sagae, Saetre, Matsuzaki, & Tsujii, 2009). IE techniques typically involve several steps such as named-entity tagging, syntactic parsing, and rule matching. These required steps to transform text are relatively expensive. Processing large text databases creates difficult challenges for IE in leveraging and extracting information from relational databases. IE techniques proposed so far are not feasible for large databases or for the web, since it is not realistic to tag and parse every available document. In addition, IE requires a dictionary of entities and relational terms or well-defined rules for identifying them. This requirement posed by IE is challenging because unstructured text corpora tend to be 1) non-uniform and incomplete, 2) synonymous and aliased, and 3) polysemous (Carpineto & Romano, 2010). These factors are attributed to low recall of IE systems reported in the literature. Other major problems with current IE techniques are that they are labour intensive in terms of processing text collections and require extensive knowledge on the target domain (Califf & Mooney, 2003). Although it is currently feasible to apply sentence parsers or named entity extraction tools to the entire PubMed database, a portability issue with doing so remains problematic particularly when it is applied to other types of datasets other than PubMed. Due to these issues, IE is not applicable to some domains where human intervention is necessary or when domain experts are not available.
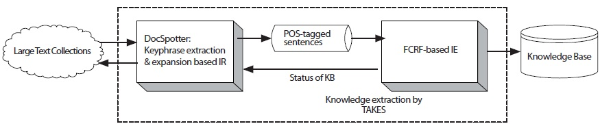
The goal of this paper is to develop an integrated knowledge extraction system, TAKES, that overcomes the aforementioned issues with current IE systems. TAKES stands for Two-step Approach for Knowledge Extraction System. The specific research objectives are 1) whether keyphrase-based query expansion improves retrieval performance and 2) whether Feature-enriched Conditional Random Field (FCRF)-based information extraction enhances the extraction accuracy. By enabling both to retrieve promising documents that contain target biological entities relations and to extract those target entities and relations, TAKES helps curators and biologists discover new entities and relations buried in a large amount of biomedical data. Figure 1 shows the overall diagram of TAKES.
Although several papers have suggested the development of a scalable knowledge extraction system (i.e., Textpresso) (Agichtein & Gravano, 2003; Banko & Etzioni, 2007; Hu & Shen, 2009; Shatkay & Feldman, 2003; Muller, Kenny, & Sternberg, 2004), the proposed technique is differentiated and unique from existing knowledge extraction systems in the following ways. First, we introduce a novel query expansion technique based on keyphrases, while others are based on a single term or are combined with a rule-based learning technique like Ripper (Cohen & Singer, 1996). Second, our system is based on unsupervised query training whereas others are based on supervised querying learning. Third, we introduce a FCRF-based extraction technique for knowledge extraction to information extraction tasks. In addition, seamlessly integrating retrieval and extraction into a knowledge extraction system is a major strength of our approach, and it is proven that TAKES is highly effective by the experimental results.
The details of TAKES are provided in Section 3. The experimental results show that two main components of TAKES, DocSpotter and FCRF, outperformed the other compared query expansion and information extraction techniques, respectively. The detailed description is provided in Section 4.
The rest of the paper is organized as follows: Section 2 proposes and describes a novel knowledge extraction technique; Section 3 explains the experimental settings and evaluation methodologies. We report and analyze the experimental results in Section 4. Section 5 concludes the paper and suggests future work.
2. APPROACH AND METHOD
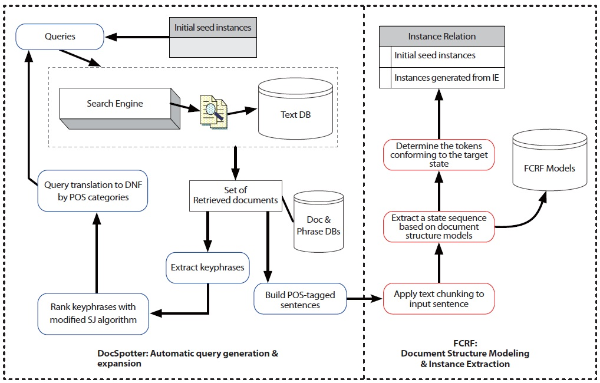
In this section, we describe the architecture of TAKES (Figure 2). Note that the components in blue boxes (DocSpotter) are described in Section 2.1.2, and the components in red boxes (FCRF) are described in Section 2.2.2. TAKES uses a pipelined architecture and extracts target entities with as little human intervention as possible.
The TAKES algorithm works using the following steps:
Step 1: Starting with a set of user-provided seed instances (the seed instance can be quite small), our system retrieves a sample of documents from the text databases. Note, an instance denotes a protein-protein interaction pair. At the initial stage of the overall document retrieval process, we have no information about the documents that might be useful for the goal of extraction. The only information we require about the target relation is a set of user-provided seed instances, including the specification of the relation attributes to be used for document retrieval. We construct some simple queries by using the attribute values of the initial seed instances to extract the document samples of a predefined size using the search engine.
Step 2: The instance set induces a binary partition (a split) on the documents: those that contain instances or those that do not contain any instance from the relation. The documents are labeled automatically as either positive or negative examples, respectively. The positive examples represent the documents that contain at least one instance. The negative examples represent documents that contain no instances.
Step 3: TAKES next applies data mining and IR techniques to derive queries targeted to match—and retrieve— additional documents similar to the positive examples.
Step 4: TAKES then applies a CRF-based state sequence extraction technique over the documents. It models a set of document structures using the training documents. These models are kept in the model base which will serve as an engine for extracting state sequence from the documents.
Step 5: The system queries the text databases using the automatically learned queries from Step 3 to retrieve a set of promising documents from the databases and then returns to Step 2. The whole procedure repeats until no new instances can be added into the relation or we reach the pre-set limit of a maximum number of text files to process.
2.1. DocSpotter
DocSpotter is a novel querying technique to iteratively retrieve promising documents, specifically tuned for information extraction. DocSpotter uses several DM and Natural Language Processing (NLP) techniques. First, DocSpotter adopts a keyphrase-based term selection technique combined with data mining and natural language processing techniques. Second, it proposes a query weighting algorithm combined with the modified Robertson Spark-Jones (RSJ) weight algorithm and the Information Gain algorithm. Third, it translates a query to the Disjunctive Normal Form (DNF) by POS (Part-Of-Speech) term categories.
2.1.1 Data Collections for DocSpotter Evaluation: Subset of MEDLINE and TREC
Two different data collections, TREC and MEDLINE, were used for the evaluation for DocSpotter.
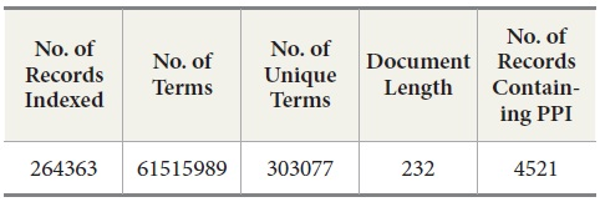
MEDLINE: We collected a subset of MEDLINE data consisting of 264,363 MEDLINE records from PubMed at http://www.ncbi.nlm.nih.gov/pubmed. These records were retrieved in the XML format by PubMed APIs provided in the Entrez E-Utilities package. After we collected the records, we queried BIND PPI DB with PubMed ID to identify what records contain PPI pairs. Out of these 264,363 records, there are 4521 records containing protein-protein interaction pairs (Table 1). Since the collected dataset does not have a sufficient number of records that contain protein-protein interaction pairs, we decide to include the records containing protein-protein pairs from other sources than human expert-curated databases such as OMIM (Online Mendelian Inheritance in Man) and DIP (Database of Interacting Proteins). The reason for this decision is that we are interested in how well DocSpotter can find the records that have not been covered in these two databases. Out of 4521 records, 4100 records were identified by OMID and DIP and the rest of the records, 421, are from other sources (He, Wang, & Li, 2009).
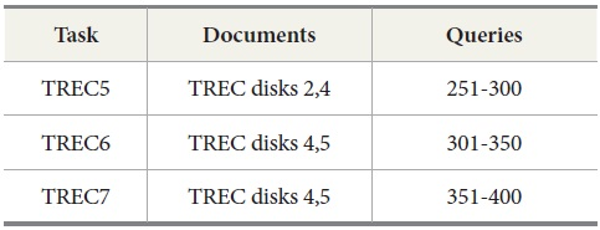
TREC: To evaluate the performance of DocSpotter on the standard IR evaluation collection, we used TREC data (TREC-5, TREC-6, and TREC-7 ad hoc test set). These TREC ad hoc test sets have been used in many different tracks of TREC including the TREC-5 routing track, the TREC-7 ad hoc track, and the TREC-8 ad hoc track. We purchased these TREC data collections from Linguistic Data Consortium (LDC). The ad hoc task investigates the performance of systems that search a static document collection using new query statements. The document set consists of approximately 628,531 documents distributed on three CD-ROM disks (TREC disks 2, 4, and 5) taken from the following sources: Federal Register (FR), Financial Times (FT), Foreign Broadcast Information Service (FBIS), LA Times (LAT), Wall Street Journal, PA Newswire, and Information from Computer Select disks.
The query sets and document collections used in these tasks are shown in Table 2. Two tasks were conducted for evaluation. The first task is to retrieve results for the query sets from 251 to 400 used in TREC 5, 6, and 7. The second task with MEDLINE data is to retrieve MEDLINE records that contain protein-protein interaction pairs with two IR systems, PubMed and Lemur, to query MEDLINE data.
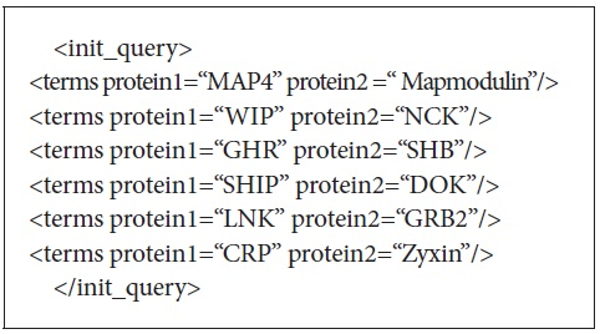
There were 25 initial queries provided for the experiments. These queries consisted of 3 to 5 protein-protein interaction pairs. Figure 3 shows the initial query used to retrieve the documents from PubMed. The initial queries were passed to either PubMed or Lemur to retrieve the initial set of retrieved documents. DocSpotter was applied to extract keyphrases and expand queries based on the top N ranked keyphrases. The performance of DocSpotter and other comparison techniques is measured by average precision, precision at rank n, and F-measure on MEDLINE and TREC-5, 6, and 7 data. The details of the performance measure are described in Section 3.
2.1.2 DocSpotter Description
The keyphrase extraction technique used for DocSpotter consists of two stages: 1) building the extraction model and 2) extracting keyphrases. Input of the “building extraction model” stage is training data whereas input of the “extracting keyphrases” stage is test data. These two stages are fully automated. Both training and test data are processed by the three components: 1) Data Cleaning, 2) Data Tokenizing, and 3) Data Discretizing. Through data cleaning and tokenizing, we generate candidate keyphrases. Three feature sets were chosen and calculated for each candidate phrase: 1) TF*IDF, 2) Distance from First Occurrence, and 3) POS Tagging. Since these features are continuous, we need to convert them into nominal forms to make our machine learning algorithm applicable. Among many discretization algorithms, we chose the equal-depth (frequency) partitioning method which allows for good data scaling. Equaldepth discretization divides the range into N intervals, each containing approximately the same number of samples. During the training process, each feature is discretized. In DocSpotter, the value of each feature is replaced by the range to which the value belongs.
Keyphrase Ranking
Automatic query expansion requires a term-selection stage. The ranked order of terms is of primary importance in that the terms that are most likely to be useful are close to the top of the list. We re-weight candidate keyphrases with an Information Gain measure. Specifically, candidate keyphrases are ranked by an Information Gain, GAIN(P), a measure of expected reduction in entropy based on the “usefulness” of an attribute A. The usefulness of an attribute is determined by the degree of uncertainty reduced when the attribute is chosen. This is one of the most popular measures of association used in data mining. For instance, Quinlan (1993) uses Information Gain for ID3 and its successor C4.5 which are widely-used decision tree techniques. ID3 and C4.5 construct simple trees by choosing at each step the splitting feature that “tells us the most” about the training data. Mathematically, Information Gain is defined as:
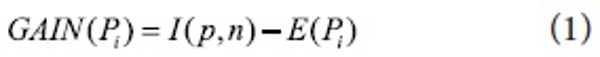
where Pi is value of candidate phrase that falls into a discrete range. I(p,n) measures the information required to classify an arbitrary tuple.
Each candidate phrase, extracted from a document, is ranked by the probability calculated with GAIN(P). In our approach, I(p,n) is stated such that the class p is where a candidate phrase is “keyphrase” and the class n is where a candidate phrase is “non-keyphrase.” Many query re-ranking algorithms are reported in literature (Robertson, Zaragoza, & Taylor, 2004). These algorithms attempt to quantify the value of candidate query expansion terms. Formulae estimate the term value based on qualitative or quantitative criteria. The qualitative arguments are concerned with the value of the particular term in retrieval. On the other hand, the quantitative argument involves some specific criteria such as a proof of performance. One example of the qualitative-based formula is the relevance weighting theory.
While there are many promising alternatives to this weighting scheme in IR literature, we chose the Robertson-Sparck Jones algorithm (Robertson & Sparck, 1976) as our base because it has been demonstrated to perform well, is naturally well suited to our task, and incorporating other term weighting schemes would require changes to our model.
The F4.5 formula proposed by Robertson and Jones has been widely used in IR systems with some modifications (Okapi). Although a few more algorithms were derived from the F4.5 formula by Robertson and Jones, in this paper we modify the original for keyphrases as shown:
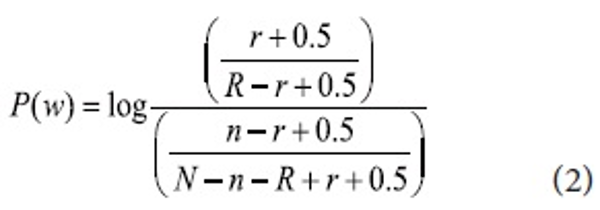
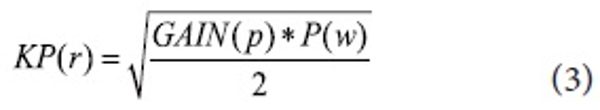
P(w) is the keyphrase weight, N is the total number of sentences, n is the number of sentences in which that query terms co-occur, R is the total number of relevant sentences, and r is the number of relevant sentences in which the query terms co-occur. We combine Information Gain with the modified F4.5 formula to incorporate keyphrase properties gained (see formula 3). All candidate keyphrases are re-weighted by KP(r) and the top N ranked keyphrases are added to the query for the next pass. The N number is determined by the size of the retrieved documents.
Query Translation into DNF
A major research issue in IR is easing the user’s role of query formulation through automating the process of query formulation. There are two essential problems to address when searching with online systems: 1) initial query formulation that expresses the user’s information need; and 2) query reformulation that constructs a new query from the results of a prior query (Abdou & Savoy, 2008). The latter effort implements the notion of relevance feedback in IR systems and is the topic of this section. An algorithm for automating Boolean query formulation was first proposed in 1970. It employs a term weighting function first described in Frants and Shapiro (1991) to decide the “importance” of terms which have been identified. The terms were aggregated into “sub-requests” and combined into a Boolean expression in disjunctive normal form (DNF). Other algorithms that have been proposed to translate a query to DNF are based on classification, decision-trees, and thesauri. Mitra, Singhal, and Buckely (1998) proposed a technique for constructing Boolean constraints.
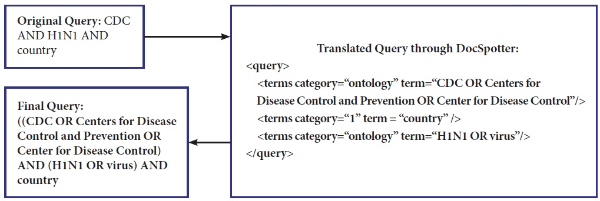
Our POS category-based translation technique differs from others in that ours is unsupervised and is easily integrated into other domains. In our technique, there are four different phrase categories defined: 1) ontology phrase category, 2) non-ontology noun phrase category, 3) non-ontology proper noun phrase category, and 4) verb phrase category. Phrases that have corresponding entities in ontologies such as MESH and WordNet belong to the ontology phrase category. Synonym relations are used for the entity matching between phrases in a query and ontologies entities. We include the verb phrase category as a major category because important verb phrases play a role in improving the retrieval performance. Keyphrases within the category are translated into DNF and categories are then translated into Conjunctive Normal Form. As explained earlier, within the same category the phrases are combined with the OR Boolean operator. Between categories, the terms are combined with the AND Boolean operator. Figure 4 illustrates how the original query (CDC and H1N1 and country) is expanded and translated into the final query with MESH. We index the MESH tree in XML, and the query term is used to look up the MESH index to select the closest match between the term and the MESH entry. Our query translation technique does not currently address the problem of translating ambiguous terms.
2.2. FCRF
In this section, we describe FCRF, a novel extraction technique based on incorporating various features into Conditional Random Fields (CRF). CRF is a discriminative undirected probabilistic graphical model to help the IE system cope with data sparseness (Lafferty, McCallum, & Pereira, 2001).
2.2.1 Data Collections for FCRF Evaluation
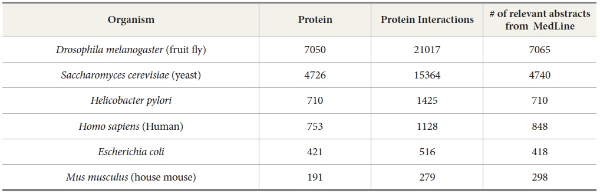
We use OMIM and DIP as references to compare the number of articles needed to be retrieved and extracted by TAKES in order to rebuild the protein-protein interactions or gene-disease interactions for each species. OMIM is a database of human genes and genetic disorders (McKusick, 1998). With OMIM, our task is to extract gene-disease interaction. For our experiment, we used the data set compiled by Ray and Craven (2001). DIP (Xenarios & Eisenberg, 2001) is a knowledge base about the biological relationships of protein-protein interactions and is constructed by human experts manually, a many-year effort. DIP, the manually curated knowledge database, serves as an ideal testbed to verify the performance of our TAKES system. It contains the information of protein names, protein-protein interaction pairs, and the MEDLINE abstracts from which the protein-protein pairs are manually extracted for a few species such as human beings, yeast, fruit flies, house mice, Helicobacter pylori, and Escherichia coli (See Table 3).
The performance of FCRF and other comparison techniques is measured by precision, recall, and F-measure on MEDLINE data combined with OMIM and DIP, The details of the performance measure are described in Section 3.
2.2.2 Feature-Enriched CRF (FCRF) Models
CRF is a probabilistic framework for labeling and segmenting sequential data. The underlying idea of CRF is that of defining a conditional probability distribution over label sequences given a particular observation sequence, rather than a joint distribution over both label and observation sequences. The advantage of CRFs over HMMs is their conditional nature, resulting in the relaxation of the independence assumptions required by HMMs in order to ensure tractable inference (Lafferty, McCallum, & Pereira, 2001). In this paper, we incorporate various features such as context, linguistic, part-of-speech, text chunking, and dictionary features into the extraction decision. Feature selection is critical to the success of machine learning approaches. We will illustrate how to calculate values of feature functions.
Entity extraction can be thought of as a sequence segmentation problem: each word is a token in a sequence to be assigned a label (e.g. PROTEIN, DNA, RNA, CELL-LINE, CELL-TYPE, or OTHER). Let o=<o1, o2, …, on> be a sequence of observed words of length n. Let S be a set of states in a finite state machine, each corresponding to a label l ∈ L (e.g. PROTEIN, DNA, etc.). Let S = <s1, s2, …, sn> be the sequence of states in S that correspond to the labels assigned to words in the input sequence o. Linear-chain CRFs define the conditional probability of a state sequence given an input sequence to be:
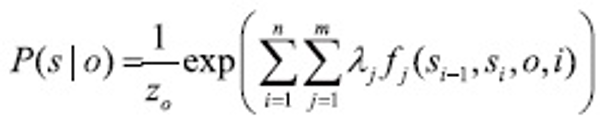
where Z0 is a normalization factor of all state sequences, fj(si-1, si, o, i) is one of m functions that describes a feature, and λj is a learned weight for each such feature function. These weights are set to maximize the conditional log likelihood of labelled sequences in a training set: D={<o,l>(1), …, <o,l>(n)}
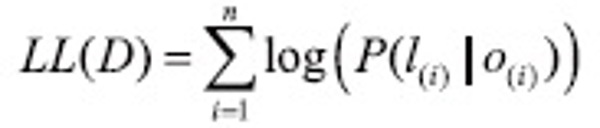
When the training state sequences are fully labelled and unambiguous, the objective function is convex, thus the model is guaranteed to find the optimal weight settings in terms of LL(D). Once these settings are found, the labelling for the unlabelled sequence can be done using a modified Viterbi algorithm. CRFs are presented in more complete detail by Lafferty et al. (2001).
Text Chunking Feature
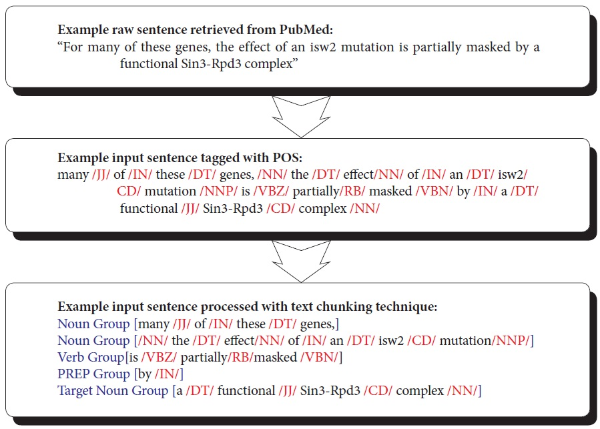
Text chunking is defined as dividing text into syntactically correlated parts of words (Kudo & Matsumoto, 2000). Chunking is a two-step process: identifying proper chunks from a sequence of tokens (such as words), and then classifying these chunks into grammatical classes. A major advantage of using text chunking over full parsing techniques is that partial parsing, such as text chunking, is much faster and more robust, yet sufficient for IE. SVM-based text chunking was reported to produce the highest accuracy in the text chunking task (Kudo & Matsumoto, 2000). The SVMbased approaches, such as the inductive-learning approach, take as input a set of training examples (given as binary valued feature vectors) and find a classification function that maps them to a class. In this paper, we use Tiny SVM (Kudo & Matsumoto, 2000) in that Tiny SVM performs well in handling a multi-class task. Figure 5 illustrates the procedure of converting a raw sentence from PubMed to the phrase-based units grouped by the SVM text-chunking technique. The top box shows a sentence that is part of abstracts retrieved from PubMed. The middle box illustrates the parsed sentence by POS taggers. The bottom box shows the final conversion made to the POS tagged sentence by the SVM-based text chunking technique.
Context Feature
Words preceding or following the target word may be useful for determining its category. It is obvious that the more context words analyzed the better and more precise the results gained. However, widening the context window rapidly increases the number of possibilities to calculate. In our experience, a suitable window size is five.
Part-Of-Speech Feature
Part of speech information is quite useful to detect named entities. Verbs and prepositions usually indicate a named entity’s boundaries, whereas nouns not found in the dictionary are usually good candidates for named entities. Our experience indicates that five is also a suitable window size. We used the Brill POS tagger to provide POS information.
Dictionary Feature
We use a dictionary feature function for every token in the corpus. This feature, described as “dictionary name” plus length of token string, informs us of whether a token matches a dictionary entry and whether it is part of a multi-token string that matches a compound named entity or named entity phrase in the dictionary.
3. EVALUATION
We evaluate TAKES on two different tasks for Information Retrieval and Information Extraction in a biomedical domain. The performance evaluation of TAKES accounts for effectiveness of each component (retrieval and extraction) to the overall evaluation. The first set of experiments aims at evaluating the performance of DocSpotter. The second set of experiments focuses on evaluating the performance of FCRF. The details of our experimental evaluation are provided in the sub-sections.
3.1. Evaluation of DocSpotter and FCRF
In this section, we explain methodologies and strategies used for evaluation of DocSpotter and FCRF. To evaluate the performance of DocSpotter, we implemented two query expansion algorithms. The first algorithm is SLIPPER which is a rule-based query expansion technique (Cohen & Singer, 1996). The second algorithm is BM25, which is a statistical expansion technique (Robertson, Zaragoza, & Taylor, 2004). These two algorithms are well-accepted query expansion algorithms (Feng, Burns, & Hovy, 2008).
In evaluation of FCRF, we do not attempt to capture every instance of such tuples. Instead, we exploit the fact that these tuples tend to appear multiple times in the types of collections that we consider. As long as we capture one instance of such a tuple, we consider our system to be successful. To evaluate this task, we adapt the recall and precision metrics used by IR to quantify the accuracy and comprehensiveness of our combined table of tuples. Our metrics for evaluating the performance of an extraction system over a collection of documents D include all the tuples that appear in the collection D.
We conduct a series of experiments. We start with a few protein-protein interaction pairs or gene-disease interaction pairs and then let TAKES automatically construct queries, select the relevant articles from MEDLINE, and extract the protein-protein interaction for each species. We repeat the experiments for each species several times with different seed instances and take the average of the articles numbers. Identifying several key algorithms proposed in IE from our literature review, we implement five IE algorithms that were reported to produce high extraction accuracy. These five algorithms are 1) Dictionary-based (Blaschke, Andrade, Ouzounis, & Valencia, 1999), 2) RAPIER (Cohen & Singer, 1996), 3) Single POS HMM (Ray & Craven, 2001), 4) SVM (Kudo & Matsumoto, 2000), and 5) Maximum Entropy (Manning & Klein, 2003).
3.2. Evaluation Measure for DocSpotter and FCRF
The retrieval effectiveness of DocSpotter was measured by precision at rank n and non-interpolated average precision. Using the precision at rank n for the IR evaluation is based on the assumption that the most relevant hits must be in the top few documents returned for a query. Relevance ranking can be measured by computing precision at different cut-off points. Precision at rank n does not measure recall. A new measure called average precision combines precision, relevance ranking, and overall recall. Average precision is the sum of the precision at each relevant hit in the hit list divided by the total number of relevant documents in the collection. The cutoff value for the number of retrieved documents is 1000 in the TREC evaluation. In the evaluation of DocSpotter, we used 200 as the cutoff value in that the collection size in our evaluation is smaller than in the TREC evaluation.
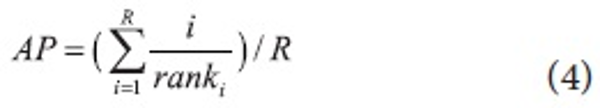
where R = number of relevant docs for that query and i/ranki = 0 if document i was not retrieved.
The extraction effectiveness of FCRF was measured by Recall, Precision, and F-measure. In IE, the evaluation of system performance is done with an answer key that contains annotations and their attributes (also called slots) that the system should find from the input. Precision (P) and recall (R) have been used regularly to measure the performance of IE as well as IR. Recall denotes the ratio of the number of slots the system found correctly to the number of slots in the answer key. In addition, since F-measure provides a useful tool for examining the relative performance of systems when one has better precision and the other better recall, we report this number where it is useful.
4. EXPERIMENTAL RESULTS AND ANALYSIS
In this section, we briefly describe the experimental settings designed to evaluate DocSpotter and FCRF and report the experimental results. The experiments are conducted to investigate two research problems studied in this paper: 1) the effectiveness of DocSpotter, a query expansion technique and 2) the effectiveness of FCRF, an information extraction technique.
4.1. Experimental Results with DocSpotter
As stated in the previous section, DocSpotter, the keyphrase-based query expansion algorithm, was evaluated with two different data sets and search engines. The subsections below report the experimental results with these combinations. We used 25 initial queries for both MEDLINE – PubMed and MEDLINE – Lemur. In order to examine whether query drift (i.e., the presence of aspects or topics not related to the query in top-retrieved documents) exists, we ran ten iterations in each query expansion experiment. Two measures, average precision and precision at top 20 documents, were utilized for performance evaluation of our query expansion algorithm. We limited the number of retrieved documents to 200. The four query expansion algorithms shown below are used for the experiments:
-
BM25: Okapi BM25 algorithm.
-
SLP: SLIPPER, a Rule-based AdaBoost algorithm
-
KP: Keyphrase-based query expansion algorithm
-
KP+C: In addition to the KP formula, this algorithm employs Boolean constraints by POS type of keyphrases and serves the key algorithm for DocSpotter.
Experimental Results of DocSpotter on TREC Data
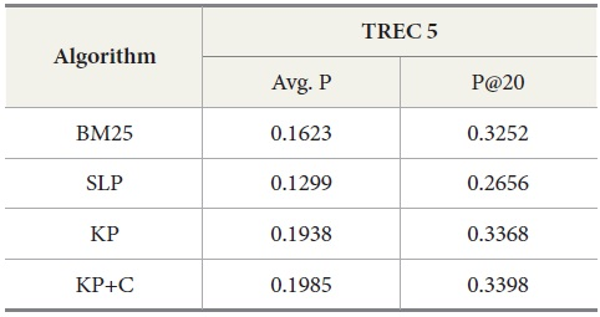
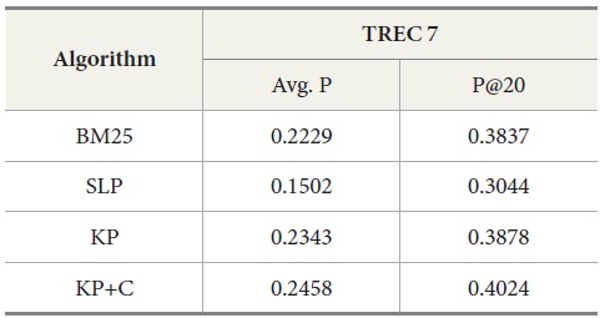
Table 4 shows the overall performance of the four algorithms executing the query set 251-300 on TREC 5 data. The results show that KP has the best performance in average precision as well as in precision at top twenty ranks (P@20) compared to other algorithms. To confirm the differences among the conditions, we conducted an ANOVA for the P@20 TREC 5 results. This showed an overall effect of condition F(3,196)=17.64, p<0.01. We also conducted individual t-tests essentially as specific comparisons. Our prediction that KP would be better than BM25 was confirmed t(49)=-7.37, p<0.01 (one-tailed) at n-1 degrees of freedom (50 queries). Similarly, our prediction that KP+C would be better than KP was confirmed t(49)=-4.72, p<0.01 (one-tailed).
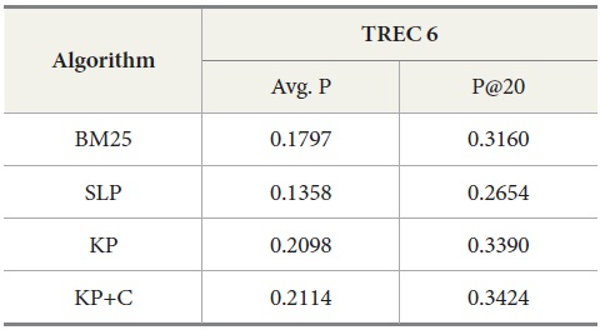
Tables 5 and 6 show similar results to those obtained for TREC 5. The three new algorithms improve the retrieval performance on TREC 6 and 7. As with TREC 5, the KP+C algorithm outperforms BM25 and SLP algorithms in average precision and in P@20.
DocSpotter, the keyphrase-based technique combined with the POS phrase category, produces the highest average precision. One of the best results on TREC 5 is 19.44 and 32.40 in average precision and P@20 respectively (Mitra, Singhal, & Buckely, 1998). On TREC 6, their best results are 20.34 and 33.50 in average precision and P@20. The algorithm KP+C produces 21% and 48% better than these results on TREC 5 in average precision and P@20. On TREC 6, it is 39% and 22% which are better than the results reported by Mitra et al. (1998).
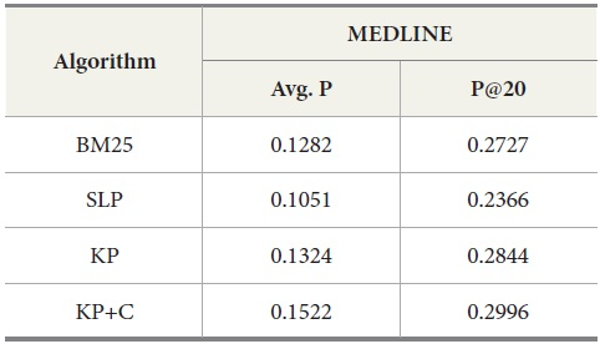
Experimental Results of DocSpotter on MEDLINE Data
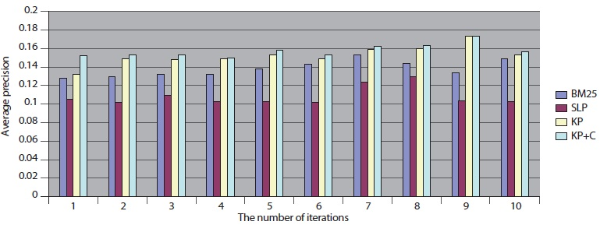
The experimental results for MEDLINE with PubMed are shown in Table 7. Our keyphrase-based technique combined with the POS phrase category produces the highest average precision. Our two algorithms (KP and KP+C) improve the retrieval performance on the tasks of retrieval documents containing protein-protein interaction pairs. The KP+C algorithm gives the best average precision. The worst performance was produced by a rule-based algorithm (SLP) both in average precision and precision at top 20.
The overall performance of the query expansion algorithms is poor in terms of average precision (Figure 6) and precision at top 20. There might be two possible reasons that cause this overall poor performance. First, PubMed is based on an exact match retrieval model which makes the keyphrase-based query expansion less effective. Second, the size of the database, which contains more than 18 million documents, is too big.
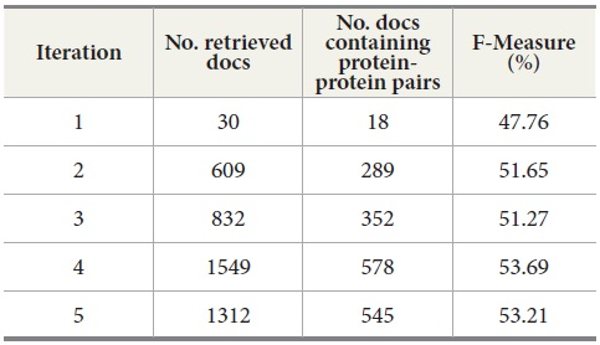
We also explored the effect of a sequence of query expansion iterations. Table 8 shows the results for five query expansion iterations. The second column shows the number of retrieved documents from MEDLINE per iteration. The third column displays the number of retrieved documents containing protein-protein pairs. The fourth column is the F-Measure. For F-Measure, we used b=2 because recall is more important than precision in the tasks of retrieving the documents containing protein-protein interaction pairs. Our results show that F-Measure generally increases as the number of iterations increases, and the results indicate that a sequence of query expansion iterations has an impact on the overall retrieval performance.
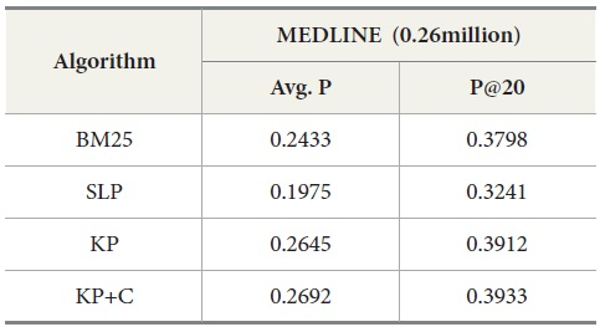
In addition, we compared DocSpotter with another query expansion algorithm on MEDLINE – Lemur. For this experiment, approximately 0.26 million MEDLINE records were indexed and searched with Lemur. As indicated in Table 9, the best performance was produced by the keyphrase-based query expansion algorithm with the POS phrase category (KP+C) both in average precision and precision at top 20. The baseline query expansion algorithm was the next highest followed by BM25.
Overall, these results with MEDLINE-Lemur are no different from the previous results. One interesting observation is that MEDLINE-Lemur produces higher scores of average precision and precision at top 20 than other dataset-search engine combinations. The reasons for these higher scores are because 1) the data collection drawn from MEDLINE is homogeneous in terms of the subject matter of the collection and 2) Lemur search engine is the better search engine to work with over PubMed.
4.2. Experimental Results with FCRF
The results of experiments to evaluate the performance of FCRF on the task of protein-protein interaction extraction are shown below. In these experiments, five machine learning algorithms were trained using the abstracts with proteins and their interactions were processed by the text chunking technique. With this set of data, these systems extracted protein-protein interactions from the retrieved documents using DocSpotter. This gave us a measure of how the protein interaction extraction systems perform alone. Performance was evaluated using ten-fold cross validation and measuring recall and precision. Since the task was to extract interacting protein-pairs, we did not consider matching the exact position and every occurrence of interacting protein-pairs within the abstract. To evaluate these systems, we constructed a precision-recall graph. Recall denotes the ratio of the number of slots the system found correctly to the number of slots in the answer key, and precision is the ratio of the number of correctly filled slots to the total number of slots the system filled.
Our experiments show that RAPIER produces relatively high precision but low recall. Similar results are observed in the Single POS HMM method which also gives high precision but low recall. MaxEnt produces the second best results, although recall is relatively lower than precision.
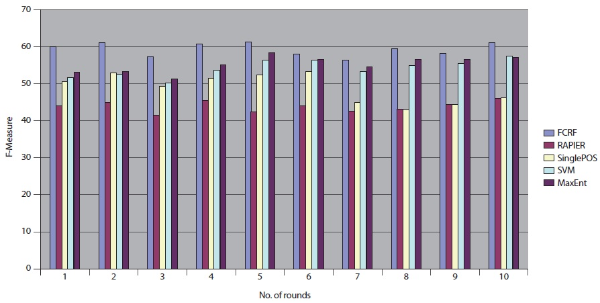
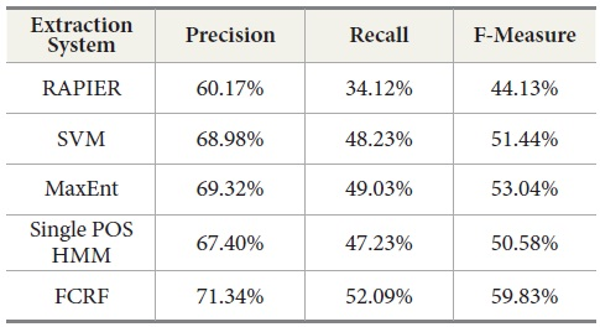
SVM produces better results than RAPIER or Single POS HMM but worse than MaxEnt and FCRF. Among these five systems, FCRF outperforms RAPIER, single POS HMM, SVM, and MaxEnt in terms of precision, recall, and F-measure. As shown in Table 10, F-Measure of FCRF is 59.83% whereas RAPER is 44.13%, SVM is 51.44%, single POS HMM is 50.58%, and MaxEnt is 53.04%.
We conducted another set of tests to investigate whether the results observed above are reproduced. To this end, we used input data that was obtained from DocSpotter as discussed. Since iterative query expansion is able to retrieve multiple sets of documents, we used a set of documents retrieved in each round.
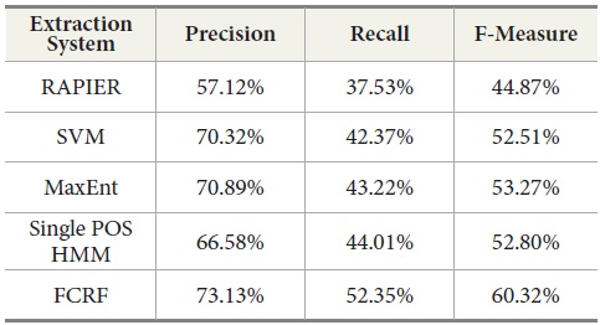
Table 11 shows the experimental results with a new set of incoming data from DocSpotter. The apparent pattern of the results resembles the one reported in the previous run (Table 10). As indicated in Table 11, FCRF produced the best performance: precision 73.13%, recall 51.91%, and F-measure, 59.36%. The next highest score is produced by MaxEnt. RAPIER produces the lowest precision, recall, and F-Measure.
We repeated the same experimental tests over the 10 different datasets that were sent from DocSpotter. Figure 7 shows the results of the five extraction methods, FCRF, Single POS HMM, RAPIER, SVM, and MaxEnt in F-Measure. FCRF outperforms the other four algorithms. FCRF produces between 56.32% and 61.12% in F-Measure. Single POS HMM produces between 42.84% and 53.32% in F-Measure. RAPIER produces between 42.23% and 45.95% in F-Measure. SVM produces between 50.20% and 57.43% in F-Measure. MaxEnt’s performance is in between 51.23% and 58.23% in F-Measure.
5. CONCLUSION AND FUTURE WORK
In this paper, we proposed a hybrid knowledge extraction algorithm drawn from several research fields such as DM, IR, and IE. Specifically, we developed a novel extraction algorithm that consists of 1) keyphrase-based query expansion to spot promising documents and 2) Feature-enriched Conditional Random Field-based information extraction. We also conducted a series of experiments to validate three research hypotheses formed in this paper.
The major contributions of this paper are threefold. First, this paper introduced a novel automatic query-based technique (DocSpotter) to retrieve articles that are promising for extraction of relations from text. It assumed only a minimal search interface to the text database, which can be adapted to new domains, databases, or target relations with minimal human effort. It automatically discovered characteristics of documents that are useful for extraction of a target relation and refined queries per iteration to select potentially useful articles from the text databases. Second, a statistical generative model, Feature-enriched Conditional Random Field (FCRF), was proposed for automatic pattern generation and instances extraction. Third, we conducted a comprehensive evaluation of TAKES with other state-of-the art algorithms. We collected 264,363 MEDLINE records from PubMed, and we also used TREC ad hoc track data collections to evaluate DocSpotter. Among MEDLINE records harvested, 4521 records contain protein-protein interaction pairs. With these records as well as OMIM and DIP protein-protein interaction databases, we evaluated the performance of FCRF. As reported in Section 4, both DocSpotter and FCRF achieved the best performance over the other comparison algorithms. As a follow-up study, we will use BioInfer, the unified PPI database, for the PPI database from which protein-protein pairs are extracted (Pyysalo, Ginter, Heimonen, Björne, Boberg, Järvinen, & Salakoski, 2007).
The results of this paper stimulate further research in several directions. First, given that key-phrasebased query expansion is proven to be effective, it is worthwhile to investigate how effective it is to apply the keyphrase-based technique to other research problems such as text summarization and categorization. Text summarization is the process of identifying salient concepts in text narrative, conceptualizing the relationships that exist among them, and generating concise representations of input text that preserve the gist of its content. Keyphrase-based text summarization would be an interesting approach to summarization in that summarizing the collections with top N ranked keyphrases generates semantically cohesive passages. In addition, a keyphrase-based approach could be applied to automatic class modeling. For example, keyphrases can be extracted from text descriptions, such as functional requirements and class model descriptions. With extracted keyphrases, we can identify a set of core classes and its relationship with other classes.
Second, it is interesting to investigate how FCRF performs when it is applied to other types of relation extractions such as subcellular-localization relation extraction. In addition, applying FCRF to other domains such as Web data extraction would be a challenging but interesting research project. In addition to relation extraction, FCRF can be applied to entity extraction such as extracting CEO names from newswires.
Third, we plan to conduct additional evaluations on other data collections such as TREC Genomics and BioCreative data. These are the standard collections that allow us to compare DocSpotter and FCRF with other state-of-the-art algorithms.