Introduction
Desertification has become a global issue, causing serious environmental and socioeconomic problems worldwide. Earth’s surface consists of 41% drylands, and approximately 25% of drylands are under threat of desertification (D’Odorico et al ., 2013; Reynolds et al ., 2007). China is one of countries most seriously affected by desertification due to rapid industrialization in the past decades (Wang et al ., 2013; Zhang & Huisingh 2018). Most desert affected lands are located in the northern part of China, with the Inner Mongolia region being one of areas most seriously damaged by desertification (Lyu et al ., 2020; Wang et al ., 2013; Wang et al ., 2020). The Chinese government has taken various measures to address and combat desertification (Zhang & Huisingh 2018).
Hulunbuir is a prefecture level city in northeastern Inner Mongolia, China. The Hulunbuir steppe, a well known steppe, is one of the four largest grasslands. It is famous as an important base of livestock husbandry in China (Nie et al ., 2005; Zhang & H uisingh 2018). Hulunbuir is also one of the four most famous sandy lands in China (Zhang & Huisingh 2018). The geographical footprint of sandy land in Hulunbuir is increasing rapidly due to fast population growth, over cultivation, over grazing of livest ock, and so on (Dong & Ya 2002; Nie et al ., 2005). In contrast, steppe coverage in Hulunbuir decreased by up to 35% in the 2000s (Park et al .,2013).
Many projects have been launched to mitigate land degradation, such as the National Action Programme (NAP), the Three‐North Shelterbelt Programme (TNSP), the Grain to Green Programme (GGP), and so on (Wang et al ., 2020; Zhao et al ., 2010). Under these policies, various studies have been conducted to increase effects of vegetation restoration (Lyu et al ., 2020). Adaptation capabilities of restored plants are important factors for the success of vegetation restoration. Tree and shrub species used for restoration have been selected as suitable species for environmental conditions of restoration regions as they ar e highly tolerant to drought, cold, and wind erosion (Wang et al ., 2020; Zhao et al ., 2007). After restoration, land enclosure is conducted using fences to prevent human disturbance and grazing of livestock, thus helping the natural reestablishment of vege tation (Gao et al ., 2002; Park et al ., 2013). Nevertheless, enclosures do not completely prevent the grazing of livestock in restoration areas for a sufficient period to allow for rehabilitation (Verdoodt et al ., 2010). Furthermore, in consideration of loc al herders’ economic interests, local governments often allow livestock grazing at certain time. Livestock grazing can produce changes in grassland attributes, including net primary production, plant species composition, and community structure (Anderson & Briske 1995). Several studies conducted in Inner Mongolia have reported preferences of grazing livestock and their effects on plant community changes (Wang et al ., 2003; Wang et al .,2014).
The objective of this study was to examine the effect of livestock grazing on restored plants at vegetation restoration sites in Hulunbuir, Inner Mongolia, China. This study sought to answer the following questions: (1) whether livestock would feed on restored plants during the growing season, (2) whether there were differences in feeding preferences according to livestock species, and (3) whether there were differences in grazing frequency according to the extent of restoration.
Materials and Methods
Study site and taxonomic survey of plants
This study was conducted from July 2014 to August 2014 in a restoration area of a grassland in Hulunbuir steppe, Inner Mongolia Autonomous Region, China (48°2312″ N, 118°1000″ E; Fig. 1, 2). The grassland is located at the boundary of semi humid and semi d ry climatic regions. The average annual temperature in Hulunbuir steppe is between -2.5°C and 0°C. The area experiences severe temperature changes of -49°C in winter and 40°C in summer. The average annual rainfall is between 270 mm and 370 mm, with more th an 70% of the rainfall occurring in July and August. Restoration areas were separated according to the year of restoration. Four restoration sites (restored in 2005, 2008, 2011, and 2012) were selected for monitoring and sampling. All restored sites were surrounded by wire entangled fences of 1.5 m high around these sites to protect planted vegetation from grazing of livestock. Part of the enclosure could only undergo grazing once or twice a year by local herders (Park et al ., 2013). The non restoration are a (NR), where sand dunes were exposed, was selected as a reference site.
Fig. 1.

Fig. 2.

Four different plants: two shrub species ( Caragana microphylla Lam. and Corethrodendron fruticosum (Pallas) B.H. Choi & H. Ohashi) and two tree species Pinus sylvestris L. var. mongolica Litv. and Populus canadensis Moench) were used for restoration by planting (restored in 2008), seeding (restored in 2005, 2012), and mixing (restored in 2011) (Fig. 3).
Fig. 3.

A taxonomic field survey was conducted for the restoration area and a non restoration area. The identification of plant species was based on the flora of China (W u et al ., 2010), the flora of grassland in China (Lu et al ., 2012), and the flora of Korea (Lee, 2003).
Sample preparation and DNA extraction
We counted the number of livestock feces detected in each restoration site and NR based on line transect surveys. Some fecal samples were collected to analyze feeding preferences of livestock. A total of 800 m transects (200 m each, 4 replicates) were randomly arranged in each restoration area. All feces of livestock were classified as either cow or horse feces according to their morphological characteristics. While collecting feces, the outer portion of feces was eliminated. Only the inner part of feces was collected and transferred individually into polyethylene bags to avoid sample contamination. All collected fecal samples were delivered to the laboratory and stored at −80°C until DNA extraction.
Genomic DNA was extracted from feces using the QIAamp® DNA Stool Mini Kit (Qiagen, USA) following the manufacturer’s protocols excluding the lysis step. To ensure sufficient homogenization during the lysis step, we added two 5 mm stainless steel beads to samples and mixed th em by shaking on a mixer mill (Retsch, Germany) at 26 Hz for 1 min (Joo et al ., 2014). Extracted DNA was eluted in 200 µL of ATE buffer and stored at -20°C until further analyses.
Polymerase chain reaction (PCR) amplification, cloning, and sequencing
We selected the internal transcribed spacer 2 (ITS2) gene region for the identification of plant species in fecal samples. Target genes were amplified using primer set ITS_S2F and S3R (Chen et al ., 2010). PCR was conducted in a 25 μl reaction volume, containing 1 µl of extracted DNA, 1× Ex Taq Buffer, 1.5 mM of MgCl2, 0.2 mM of dNTPs, 0.2 µM of each primer, and 1 U of Ex Taq DNA polymerase (Takara, Japan). PCR was performed in a thermal cycler (Applied Biosystems 2720, USA) using the following conditions: a n initial denaturation at 94°C for 5 min, 45 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, elongation at 72°C for 45 s, and a final extension step at 72°C for 10 min. PCR products were purified using an AccuPrep® PCR Purification Kit (Bioneer, Korea). Purified PCR products were ligated into the pGEM® T Easy Vector (Promega, USA) according to the manufacturer's protocols and transformed into DH5α chemically competent cells. Cells were plated in Luria-Bertani agar + ampicillin medium wi th X-gal solution (2% w/v) for antibiotic selection and blue white screening. After the cloning step, white colonies were selected and amplified using M13F and M13R primers for colony PCR. PCR conditions were as follows: an initial denaturation at 95°C for 10 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, elongation at 72°C for 1 min, and a final extension step at 72°C for 7 min. Sequencing was conducted using a commercial sequencing service company (Genotech, Korea). Each DNA sequence was identified using BLASTN searches of the GenBank database. Sequence alignments and length calculations were conducted using the MEGA 5 program (Tamura et al ., 2011).
Diversity of plant species detected in feces
The diversity of plant species detected in feces was determined using both the Shannon Wiener diversity index (H´) and Simpson’s index (D) as follows:


where n was the total number of each species, and N was the total number of all species (Shannon Weaver, 1949 Simpson, 1949). Simpson’s index was transformed to 1-D, with values ranging from 0 to 1. A diversity index was derived for the diet detected from feces using molecular approaches.
Statistical analysis
Analysis of variance (ANOVA) was used to test statistically significant differences in the density of livestock feces detected at each study site, and post hoc tests were performed using Tukey’s method ( P < 0.05). Fisher's exact test was used to assess differences in feeding frequency between livestock species. All statistical analyses were performed with S-Plus 8 for Windows (Insightful Corp., USA) and R program (R Core Team, 2013).
Results
Flora of study sites
A total of 41 plants were identified at the study sites. Aside from restored plants, main native plants growing in the restoration sites were Corispermum squarrosum (L.) Moq., Corispermum hyssopifolium L., Astragalus adsurgens Pall., Artemisia desertorum Spreng., Orobanche coerulescens Stephan ex Willd., Oxytropis racemosa Turcz., Agropyron cristatum (L.) Gaertn., Echinops gmelini Turcz., and so on (Fig. 3). The largest number of plant species was observed at the 2008 restoration site. The number of plant species tended to increase in areas restored long time ago (7 species in NR, 10 species in 2012, 11 species in 2011, and 19 species in 2005; Table 1).
Table 1.
| No. | Species | 2005 | 2008 | 2011 | 2012 | NR |
|---|---|---|---|---|---|---|
| 1 | Agropyron cristatum (L.) Gaertn. | + | ||||
| 2 | Allium condensatum Turcz. | + | + | |||
| 3 | Artemisia argyi H. Lév. & Vaniot | + | ||||
| 4 | Artemisia capillaris Thunb. | + | ||||
| 5 | Artemisia desertorum Spreng. | + | + | + | + | |
| 6 | Arundinella sp.1 | + | ||||
| 7 | Astragalus adsurgens Pall. | + | + | + | + | + |
| 8 | Calamagrostis pseudophragmites (Haller f.) Koeler | + | ||||
| 9 | Caragana microphylla Lam. | + | + | + | + | + |
| 10 | Carex duriuscula C.A. Mey. | + | ||||
| 11 | Chenopodium ficifolium Sm. | + | ||||
| 12 | Cleistogenes squarrosa (Trin.) Keng | + | ||||
| 13 | Corispermum hyssopifolium L. | + | + | + | + | |
| 14 | Corispermum squarrosum (L.) Moq. | + | + | + | ||
| 15 | Cuscuta australis R. Br. | + | ||||
| 16 | Echinops gmelini Turcz. | + | + | + | ||
| 17 | Epilobium sp.1 | + | ||||
| 18 | Euphorbia humifusa Willd. Hedysarum fruticosum Pall. | + | ||||
| 19 | (Corethrodendron fruticosum (Pallas) B.H. Choi & H. Ohashi) | + | + | + | + | + |
| 20 | Inula britanica L. | + | ||||
| 21 | Juncus gracillimus (Bunchenau) V. Krecz. & Gontsch. | + | ||||
| 22 | Lappula myosotis V. Wolf | + | ||||
| 23 | Leymus secalinus (Georgi) Tzvelev | + | ||||
| 24 | Medicago polymorpha L. | + | ||||
| 25 | Olgaea lomonosowii (Trautv.) Iljin. | + | ||||
| 26 | Orobanche coerulescens Stephan ex Willd. | + | + | + | ||
| 27 | Oxytropis racemosa Turcz. | + | + | + | + | |
| 28 | Phragmites australis (Cav.) Trin. ex Steud. | + | ||||
| 29 | Pinus sylvestris L. var. mongolica Litv. | + | ||||
| 30 | Plantago sp.1 | + | ||||
| 31 | Populus canadensis Moench | + | + | |||
| 32 | Potentilla acaulis L. | + | ||||
| 33 | Salix cheilophila C.K. Schneid. | + | ||||
| 34 | Setaria viridis (L.) P. Beauv. | + | + | + | ||
| 35 | Silene jenisseensis Willd. | + | ||||
| 36 | Sonchus arvensis L. | + | ||||
| 37 | Sophora flavescens Aiton | + | ||||
| 38 | Suaeda glauca (Bunge) Bunge | + | ||||
| 39 | Taraxacum asiaticum Dahlst. | + | ||||
| 40 | Thymus mongolicus (Ronniger) Ronn. | + | ||||
| 41 | Ulmus pumila L. | + | + |
Species diversity detected in feces and the mean fecal density of livestock
A total of 102 livestock feces samples were collected from the four different restoration areas and non restoration sites (Table 2). The species diversity index of plants detected in livestock feces differed according to the study site (H’: 1.88-2.18, 1-D : 0.79-0.90, Table 2). The restoration site with the highest species diversity was the 2008 site ( H´ 2.48, 1-D : 0.90), while the site with the lowest species diversity was the 2005 site (H´ : 1.88, 1-D : 0.79). There was no significant difference in the species diversity of food sources by livestock species, although more plant species were detected in cow feces (Table 2).
Table 2.
| No. of feces | Amplification success (%) | Shannon index (H') | Simpson's index (1-D) | |
|---|---|---|---|---|
| Study site | ||||
| 2005 | 33 | 20(60.6) | 1.88 | 0.79 |
| 2008 | 24 | 19(79.2) | 2.48 | 0.90 |
| 2011 | 15 | 13(86.7) | 2.14 | 0.86 |
| 2012 | 12 | 9(75.0) | 1.89 | 0.82 |
| NR (non-restored) | 18 | 16(88.9) | 2.18 | 0.86 |
| Livestock | ||||
| Cow | 38 | 27(71.1) | 2.07 | 0.79 |
| Horse | 64 | 50(78.1) | 2.79 | 0.92 |
| Total | 102 | 77(75.5) | 1.44 | 0.91 |
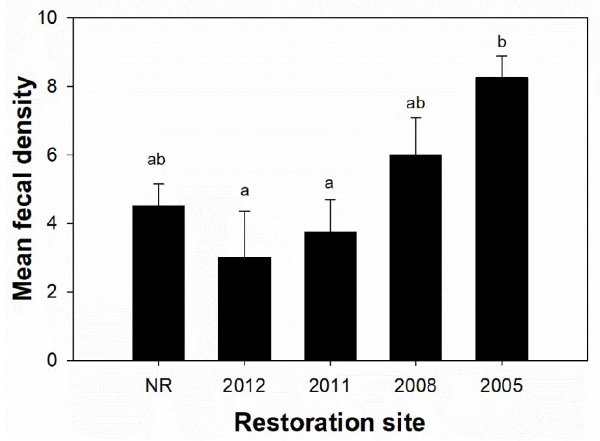
Mean (± SE) fecal densities ranged from 3.0 ± 1.4 feces/replicate at the 2012 site to 8.3 ± 0.6 feces/replicate at the 2005 site (Fig. 4). The mean fecal density at NR sites was 4.5 ± 0.65, which was higher than those at the two restored areas in recent years. The mea n fecal density was positively correlated with the restoration period. It was significantly different between the 2005 site and the 2011 or 2012 site ( P < 0.05) (Fig.4).
Fig. 4.
Feeding preference of livestock
We detected plant species genes from 77 (75.5%) out of 102 fecal samples collected from the study sites. The amplification success rate ranged from 60.6% to 88.9%, depending on the year of restoration. It showed no significant difference between livestock species (Table 2).
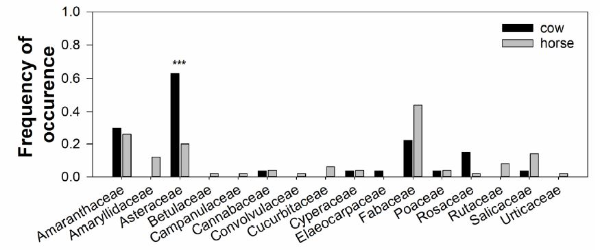
A total of 35 different sequences were detected via DNA barcoding. Plant species were identified at the species or genus level by comparison with GenBank (Table 3). Most sequences showed similarities of more than 98%. A total of 17 potential plant species were detected in comparison with results of the vegetation survey at the study sites (Table 3). Two shrub species, C. microphylla Lam. and C. fruticosum Pall., were detected in the feces of livestock. However, Caragana sp. was only detected at a low frequency (< 0.2) in livestock feces collected at the 2008 restoration site. In contrast, Corethrodendron sp. was detected with a frequency from 0.05 to 0.25 in most restored and non restored sites, excluding the 2012 site (Fig. 5). In most restoration sites, three families, Amaranthaceae, Asterceae, and Fabaceae, comprised a large portion of total detected plant species. However, plant species belonging to family Amaranthaceae were not detected at the 2012 restoration site. In the case of pla nt species belonging to family Fabaceae, the frequency of occurrence was lower (0.05) in the 2005 restoration site than in other restoration sites (0.22-0.47). In particular, Fabaceae showed the highest frequency (0.69) in the NR site. The frequency of pla nt species detected in livestock feces was similar for horses and cattle, although plant species belonging to Asteraceae were detected at significantly higher rates in cow feces than in horse feces (P = 0.00053, Fig. 6).
Table 3.
| No | BLAST results | Occurrence of identified taxon on Hulunbuir | ||||
|---|---|---|---|---|---|---|
| Description | Query coverage | E value | Identities | Accession | ||
| 1 | Allium anisopodium | 100% | 2E-169 | 99% | KF143822 | A. condensatum Turcz. |
| 2 | Allium sp. | 100% | 1E-165 | 99% | GQ412199 | |
| 3 | Allium splendens | 100% | 2E-169 | 99% | GQ412241 | |
| 4 | Amaranthus retroflexus | 100% | 7E-167 | 100% | KF493799 | |
| 5 | Ambrosia trifida | 100% | 7E-167 | 100% | DQ005970 | |
| 6 | Artemisia annua | 99% | 2E-159 | 98% | KF866382 | A. desertorum Spreng. |
| 7 | Artemisia japonica | 100% | 2E-169 | 99% | GU724289 | A. argyi H. Lèv. & Vaniot |
| 8 | Artemisia rupestris | 100% | 9E-168 | 99% | AJ297261 | A. capillaris Thunb. |
| 9 | Artemisia sericea | 100% | 9E-163 | 98% | EF577290 | |
| 10 | Astragalus sp. | 100% | 9E-168 | 99% | KJ999345 | A. adsurgens Pall. |
| 11 | Bassia dasyphylla | 100% | 4E-171 | 100% | AY489195 | |
| 12 | Betula sp. | 100% | 9E-168 | 99% | JN998976 | |
| 13 | Boehmeria sp. | 100% | 4E-171 | 100% | KP093179 | |
| 14 | Caragana sp. | 100% | 7E-169 | 99% | AB262534 | C. microphylla Lam. |
| 15 | Carex gynocrates | 100% | 9E-158 | 98% | JN999047 | C. duriuscula C.A. Mey. |
| 16 | Carex maritima | 100% | 4E-166 | 99% | JN999057 | |
| 17 | Chenopodium album | 100% | 2E-154 | 97% | FN561549 |
C. ficifolium Sm. C.acuminatum Willd. |
| 18 | Citrus sp. | 100% | 1E-164 | 99% | JQ990174 | |
| 19 | Corethrodendron multijugum | 91% | 5E-155 | 100% | AB854479 | C. fruticosum Pall. |
| 20 | Corispermum sp. | 95% | 3E-162 | 100% | JF792752 |
C. hyssopifolium L. C. squarrosum (L.) Moq. |
| 21 | Cucumis sp. | 100% | 2E-169 | 99% | KF963130 | |
| 22 | Elaeocarpus sp. | 94% | 7E-154 | 98% | DQ448689 | |
| 23 | Humulus sp. | 100% | 3E-165 | 99% | KC539581 | |
| 24 | Ipomoea trifida | 100% | 4E-164 | 99% | KC621852 | |
| 25 | Luffa sp. | 96% | 3E-162 | 99% | KF487334 | |
| 26 | Oxytropis sp. | 100% | 4E-171 | 100% | KJ143722 | O. racemosa Turcz. |
| 27 | Phaseolus sp. | 100% | 7E-169 | 99% | JN617200 | |
| 28 | Platycodon grandiflorus | 100% | 3E-165 | 99% | KP058319 | |
| 29 | Puccinellia sp. | 100% | 9E-163 | 98% | KJ598984 | |
| 30 | Rubus crataegifolius | 100% | 9E-153 | 97% | GU980782 | |
| 31 | Salix sp. | 100% | 4E-164 | 99% | KM978952 | S. cheilophila C.K. Schneid. |
| 32 | Salsola sp. | 89% | 9E-148 | 99% | HM131659 | S. collina Pall. |
| 33 | Setaria italica | 100% | 2E-169 | 99% | KF012851 | S. viridis (L.) P. Beauv. |
| 34 | Suaeda corniculata | 94% | 4E-161 | 100% | FJ449820 | S. glauca (Bunge) Bunge |
| 35 | Suaeda maritima | 100% | 2E-164 | 99% | KF866386 | |
Discussion
For successful restoration, it is necessary to select appropriate species that are well adapted to local ecological environment conditions. It is also important to meet socioeconomic needs of local residents (Sacande et al ., 2020). In Inner Mongolia, animal husbandry plays an important role in rural economies. The density of grazing livestock in Inner Mongolia is much higher than in other regions of the Mongolian Plateau (Angerer et al ., 2008). In particular, in Inner Mongolia, socioeconomic factors such as total population, gross domestic product, fiscal revenue, production values, cropland area, and number of livestock affect the desertification more than climate factors (Liu et al ., 2020). Fecal analysis of livestock in this study showed that livestock fed on C. microphylla Lam. and C. fruticosum, Pall., two of the four pl ant species used for restoration in Hulunbuir. However, they did not prefer them in the summer growing season. Most plant species detected in livestock feces were species found near the restoration site or naturally introduced by vegetation recovery. In addition, there were no significant differences in livestock preference among plant species except for family Asteraceae, although we did find some differences in diet compositions detected in livestock feces depending on the restoration site. Our results su ggest that livestock grazing in the restoration areas during the summer growth period might not have a significant impact on the survival of restored plants.
Most previous studies have focused on restoration of degraded ecosystems and harsh environments via vegetation recovery in desertified areas (Liao et al ., 2019). The selection of restored plants is often focused on plants’ adaptability to climate and their ability to improve soil microbial communities (Yan & Feng 2020). Shrub species are commonly planted to combat desertification. Legumes are potential nurse plants that can improve the survival and growth of other species due to their ability to fix nitrogen (Ren et al ., 2008; Reynolds et al ., 1999; Zhao et al ., 2 007). Unpalatable native species that could be planted in heavily grazed sites are recommended as nurse plants as they can help create microhabitats for the rehabilitation of other plants (Ren et al.al., 2008; Smit et al ., 2006). Two shrub species, C. microphylla and C. fruticosum, were classified as unpalatable or less palatable species in this study. They might be helpful in increasing the survival and growth of other plants in restoration areas. We noted that at the 2008 site, the number of plant species in creased to 26 species during the enclosure periods despite the grazing of livestock in restoration sites.
The Chinese government has implemented several policies and programs to restore grassland ecosystem functions by regulating and controlling grazing pressure, including enforcing partial or total rests from grazing for several years (Zhang et al ., 2015). Previous studies have reported that enclosures have positive effects on vegetation recovery and succession in an enclosed area of a grassland where desertification is in progress (Park et al ., 2013). However, enclosure may not be suitable for severely degraded grasslands (Jones & Carter 2016; Zhang et al ., 2015). For successful restoration, it is necessary to consider both the improvement of soil fertility and the economic demands of local herders (Anderson & Briske 1995). In addition, it would be desirable to set a value of herbage mass that needs to be reached before grazing is allowed, rather than setting a time limit for rests (Zhang et al ., 2015). Our results suggest that the four plant species used for restoration in Hulunbuir steppe are less affected by livestock grazing during the summer growing season and that the current restoration method may be positive and suitable for the initial stage of restoration in Hulunbuir.
Acknowledgments
This research was conducted as part of joint research activities for Dust and Sandstorm (DSS) under the Tripartite Environment Ministers Meeting (TEMM) among Korea, China, and Japan. This research was supported by a research project (NIE 2014 0024) of the National Institute of Ecology funded by the Ministry of Environment, Korea.