Introduction
Coccinella septempunctata, known as the seven-pointed ladybug, is a natural enemy for many pests. It is distributed across various habitats worldwide and is widely used for biological insect control in various crop systems. Both the larvae and adults feed on insects of Aphidoidea, Psylloidea, and Coccoidea superfamilies, which are found in the leaves and stems of various plants (Dolling 1991; Kalushkov & Hodek, Yu et al., 2014; Zhang et al ., 2011). C. septempunctata species are easy to breed and are ideal non target organisms for studying potential toxicity, such as, Bacillus thuringiensis or RNA interference (RNAi) based living modified organisms (Alvarez Alfageme et al ., 2012 ; Harwood et al ., 2005 Harwood et al ., 2007).
Living modified (LM) organisms (RNAi gene silencing using double stranded RNA (dsRNA)) is a powerful tool for targeted gene silencing in insects and offers a novel approach for insect control. Currently, RNAi technology is used as a biotechnological tool for the analysis of gene functions in various organism s and for pest control (Burand & Hunter, 2013; Fire et al ., 1998; Katoch et al ., 2013; Zhang et al ., 2017). Insect toxicity assessment using environmental or dietary methods can be performed using sprays, bait, LM microorganisms, or LM plants (Fischer et al ., 2016; Le, 2015 Li u et al ., 2019; Pang & Mao 1979; Zhang et al ., Zhu et al ., 2011; Zhu & Palli 2019; Zotti et al ., 2018 ). However, the success of the RNAi technique depends on the effectiveness of the in vivo action of the dsRNA in the insect species. RNAi efficacy may vary depending on the expression of the target gene, dsRNA degrading enzymes in the insect, concentration of dsRNA, and the method of delivery. According to previous reports, RNAi techniques have been used to treat western corn rootworms Diabrotica virgifera virgifera ), red powder beetles Tribolium castaneum ), Colorado potato beetles Leptinotarsa decemlineata ), and two spotted ladybugs ( Adalia bipunctata ) (Baum et al ., 2007; Haller et al ., 2019 ; Yoon et al ., 2018 ). The western corn rootworm, which impacts corn yield, responds to a dsRNA diet. Several target genes of dsRNA in western corn rootworms have been evaluated. When the dsRNA encoded an essential functional protein, western corn rootworms larvae were report edly killed at low concentrations (Baum et al ., 2007). The RNAi target gene was found to be a homolog of yeast Snf7. LM crops which produce dsRNA targeting the D. virgifera Snf7 showed insect resistance by killing western corn rootworms larvae.
The class E vacuolar sorting protein Snf7 gene is present in various organisms as Vps32, CeVps32.2, hSnf7, and At2g19830 (Kim et al ., 2011; Peck et al ., 2004; Tu et al ., 1993; Winter Hauser, 2006). Using the Snf7 RNAi system, Snf7 was confirmed to play a role in several cellular processes in mammals and nematodes (Kim et al ., 2011; Lee et al. al., 2007; Ramaseshadri et al ., 2013 ; Sweeney et al ., 2006 ). Two methods have been applied to control pests using active Snf7 dsRNA molecule. The first is the use of LM plant techniques to produce active Snf7 dsRNA in plants, such as, in LM maize (MON87411). These LM maize plants produce Snf7 dsRNA targeting the Snf7 protein, leading to increased western corn rootworms larval mortality and reduced root damage (Bachman et al ., 2020 ; Bolognesi et al ., 2012). The second method is to treat crops externally with Snf7 dsRNA, for example through trunk injections, food bait, microorganism delivery systems, or topical sprays (Hunter et al ., 2012; Kunte et al ., 2020; Li et al ., 2015; Niu et al ., 2018; Romeis & Widmer 2020 ; San Miguel & Scott, 2016; Vogel et al ., 2019 ; Zhang et al ., 2010; Zhou et al ., 2008). Although target pest can be removed using these methods, we must consider the potential risk of damage to other species. LM crops made with RNAi technology are imported with domestic import approval. These might be unintentionally released during transportation and might affect domestic species. Research on the effects of RNAi based LMOs imported into South Korea on domestic species is insufficient. In particular, the risk assessments of Snf7 dsRNA molecules in domestic natural enemies, such as, ladybugs, have rarely been conducted in South Korea.
In this study, we developed a risk assessment method for C. septempunctata using Snf7 dsRNA molecules. To identify the species of ladybug before the Snf7 dsRNA risk assessment, we compared the phenotypes of three species collected in South Korea (Harmonia axyridis, C. septempunctata, and Cryptolaemus montruzieri). The 12S rRNA gene specific primer of C. septempunctata clearly differentiated C. septempunctata at the gene level. To determine whether assessing the risk to C. septempunctata using Snf7 dsRNA treatment is feasible, treatment methods and checklists for eac h growth period were specified. A large amount of Snf7 dsRNA was isolated for the risk assessment and its purity was confirmed. C. septempunctata larvae were treated with Snf7 dsRNA for 1 5 days to confirm its presence in vivo, and the risk of Snf7 dsRNA w as determined by measuring the mortality, growth, and dry weight of C. septempunctata
Materials and Methods
Insects
C. septempunctata, H. axyridis, and C. montruzieri were obtained from the Rural Development Administration (Jeonju, South Korea). Ladybu g larvae and adults were bred in a growth chamber at 23 ± 0.5 °C, with a 16 h light and 8 h dark photoperiod and 50 70% relative humidity. The ladybugs fed on Ephestia kuehniella eggs. When the number of C. septempunctata required for the test was attained, first instar larvae were used for the risk assessment 1 day after hatching.
Identification of species
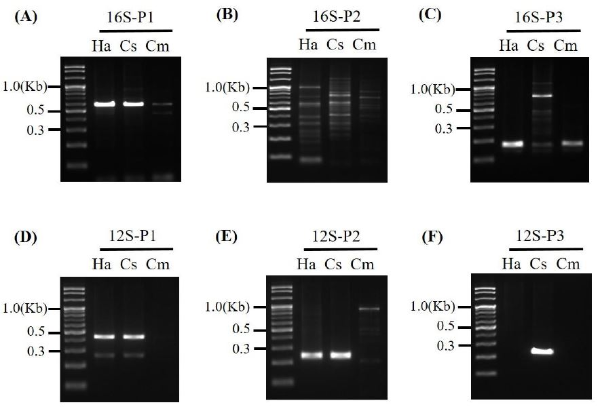
To analyze the morphological characteristics of the three ladybug species, we prepared three specimens, and observed and compared their phenotypes using a dissecting microscope (Olympus SZX16, Japan). For identification of C. septempunctata, we used two molecular marker genes, namely, 16S rRNA and 12S rRNA (Yao et al ., 2011) (Table 1). The genomic DNA of C. septempunctata, H. axyridis, and C. montruzieri was extracted using an Animal Tissues Genomic DNA Extraction Kit (Tianlong, China). The PCR products of the 16S rRNA and 12S rRNA genes in C. septempunctata were analyzed using the National Center for Biotechnology Information Basic Local Alignment Search Tool.
Table 1.
| Primer Name | Sequence (5′ 3′) | Product Size (bp) |
|---|---|---|
| 16S P1 F | CCGGTCTGAACTCAGATCACGT | 682 |
| 16S P1 R | CGCCTGTTTAACAAAAACAT | |
| 16S P2 F | TCTTCGCCTGTTTAACAAAAACATCTCTTTTT | 529 |
| 16S P2 R | GTTTTGTGGGGTGGCGCGAAGGGTATTGCCAA | |
| 16S P3 F | AAATTTGATTGGGGTGATAAAAA | 164 |
| 16S P3 R | TCGAGGTCGCAATCTTTTCT | |
| 12S P1 F | TACTATGTTACGACTTAT | 525 |
| 12S P1 R | AAACTAGGATTAGATACCC | |
| 12S P2 F | CGGGCGATGTGTACATATTTT | 227 |
| 12S P2 R | AGCAATTTTTATATCGTCGTTTTT | |
| 12S P3 F | CTTTCAAATCCAATTTCATTCTAAT | 211 |
| 12S P3 R | GTTCTGTAATTGATAATCCACGATTG |
Cloning, sequencing and dsRNA synthesis of Snf7, GFP, and vATPase A
Of the complete Snf7 mRNA sequence (968 bp), only 240 bp were used to effectively produce insecticide effects. Snf7 genes were cloned into the in vitro dsRNA expressing L4440 vector. Based on the target gene sequence of the Snf7 plasmid, Snf7 dsRNA was synthesized using the T7 promoter and the RNA polymerase T7 MEGAscript kit (Ambion, Austin, USA) (Table 2). The synthesized Snf7 dsRNA was quantified using a ND 2000 spectrophotometer (Thermo Scientific, USA) and stored in an ultra low temperature freezer at −80 °C before use. GFP and vATPase A dsRNAs were synthesized in the same manner and were used as negative and positive controls, respectively.
Table 2.
| Primer Name | Sequence (5’ 3’) | Product Size (bp) |
|---|---|---|
| DvSnf7 T7 F | TAATACGACTCACTATAGGGAGAATCCATGATATCGTGAACATC | 240 |
| DvSnf7 T7 R | TAATACGACTCACTATAGGGAGAGCAAAGAAAAATGCGTCGA | |
| GFP T7 F | TAATACGACTCACTATAGGGAGAATGGTAGATCTGACTAGTAAA | 240 |
| GFP T7 R | TAATACGACTCACTATAGGGAGAATCTGGGTATCTTGAAAAGCA | |
| vATPase A T7 F | TAATACGACTCACTATAGGGAGAGCAGAACCAGGAAGTTACAC | 700 |
| vATPase A T7 R | TAATACGACTCACTATAGGGAGATCGTAGAAGGAGGCGAGACG |
dsRNA expression analysis using RT-PCR
A total of 2 μg/μL Snf7, GFP, or vATPase A dsRNA was mixed into the artificial diet and administered to C. septempunctata larvae from each experimental group at the same time each day. After dsRNA treatment, three larvae per group were collected on day 1, 3, and 5 and immediately frozen in liquid nitrogen. Their RNAs were then transferred into RNA Stabilization Reagent (Ambion, Thermo Fisher Scientific) until RNA extraction. Reverse transcription (RT) PCR primers were designed using the Primer 3 program (https://bioinfo.ut.ee/primer3 0.4.0) (Table 3) and synthesized by Macrogen Inc. (Seoul, Republic of Korea). RT PCR was performed by synthesizing cDNA from 500 ng/μL RNA. Tubulin was used as the PCR control. mRNA expression levels of Snf7, GFP, and vATPase A in the C. septempunctata were detected by quantitative RT PCR (Applied Biosystems, Waltham, MA, USA). The 25 μL reaction mix contained 19.5 μL of water, 0.5 μL of 2.5 mM dNTP mix, 0.5 μL of each forward and reverse primers (both 10 pmole), 2.5 μL of 10× Ex buffer, 0.5 μL of Taq polymerase, and 1 μL of template. Analyses were performed using a Proplex PCR system under the following cycle conditions: initial denaturation at 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 1 min, annealing at 60 °C for 0.5 min, and extension at 72 °C for 2 min; 1 cycle of final extension at 72 °C fo r 10 min. A 5 µL aliquot of each PCR product was analyzed using gel electrophoresis on a 2.5% (w/v) agarose gel at 135 V for 25 min and the images were captured using Chemi-DocTM XRS+ (Bio Rad, Hercules, CA, USA) (Lim et al ., 2017). Three replicates were performed.
Table 3.
| Primer Name | Sequence (5’ 3’) | Product Size (bp) |
|---|---|---|
| DvSnf7 RT F | GTATTTGTGCTAGCTCCTTCGA | 120 |
| DvSnf7 RT R | TGCACTCCAAGCCCTCAAAA | |
| GFP RT F | AGAGGGTGAAGGTGATGCAA | 83 |
| GFP RT R | TTGGCCACGGAACAGGTAG | |
| VATPase A RT F TATGTTGCAAGTGTGGCC | TATGTTGCAAGTGTGGCC | 88 |
| VATPase A RT R | AACTCTCTGTCCGGTGAG | |
| Tubulin F | TTGGCCGACCAATGTACT | 117 |
| Tubulin R | CTTTCCATAGTCGACGGA |
To assess the risk of Snf7 dsRNA against C. septempunctata, we treated the larvae with high concentrations of dsRNA (2 μg/μL). The concentration was selected with reference to prior experiments (Haller et al ., 2019 ; Liang et al ., 2019; Lu et al ., Pan et al ., 2020). One larva per one petri dish was treated with Snf7, GFP, or vATPase A dsRNA (2 μg/μL) mixed (1:1) in 50% sucrose. The experiment was carried out in three replicates of three larvae (one larva per petri dish). The control group was treated with on ly 0.5 M sucrose. Snf7 dsRNA risk was evaluated based on the survival rate, development time, and dry weight of the larvae. All experiments were repeated three times. Analysis of variance was used for statistical analysis.
Results and Discussion
Identification of C. septempunctata using morphospecies and molecular markers
LM crops which produce dsRNA targeting D. virgifera Snf7 showed insect resistance by killing western corn root-worms larvae (Bachman et al ., 2013 ; Ramaseshadri et al ., 2013). The domestic natural pest enemy, C. septempunctata, is an important species for risk assessment because of the possibility of Snf7 gene transfer from herbivorous insects (aphids), which infect LM crops, to their predators. Therefore, we first identified the morphological features which can be used to identify C. septempunctata collected for Snf7 dsRNA risk assessment. Three species of ladybugs were collected and insect specimens were prepared and compared (Fig. 1). Unlike the other two ladybug species (H. axyridis and C. montruzieri), the wing cases of C. septempunctata were red with three spots clearly visible on each side. There was also one spot on the boundary of the wing, resulting to a total of seven spots. The results confirmed that the 12S-3 rRNA (211 bp) primer specifically amplified the C. septempunctata gene (Fig. 2F). In addition, nucleotide sequence analysis of the 12S-3 rRNA PCR product revealed the sequence of 12S rRNA derived from C. septempunctata (data not shown). Based on these results, we devised a method to identify C. septempunctata using morphological and molecular markers for LMO risk assessment in South Korea.
Fig. 1.

Fig. 2.
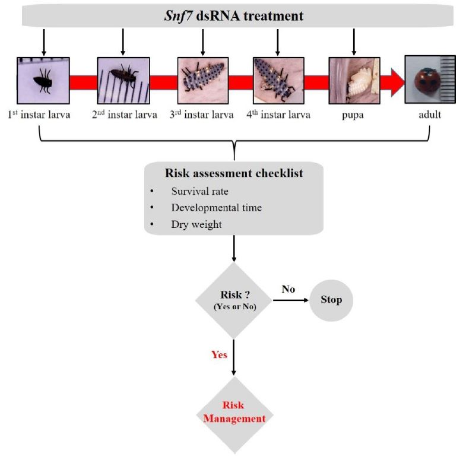
Establishment of risk assessment method using Snf7 dsRNA
To conduct risk assessments for selected domestic insect species, specific risk assessment methods were developed based on the growth characteristics of each species. Therefore, we devised the following dietary risk assessment to determine the risk of Snf7 dsRNA in C. septempunctata (Fig. 3). Firstly, the ecology of C. septempunctata from the larval to adult stages was identified. Then, the first instar larvae that hatched from the eggs were treated with Snf7 dsRNA until adulthood. The risk assessment checklist for C. septempunctata relied on survival rate, developmental time, and dry weight. If there are no differences in the risk assessment checklists between treated and control groups, the risk assessment could be stopped. However, if a potential risk was identified, risk assessment using LM crops was carried out for risk management. This risk assessment method could be applied in various ways depending on the domestic risk assessment species and the LMO expression products, such as, dsRNA and proteins.
Fig. 3.
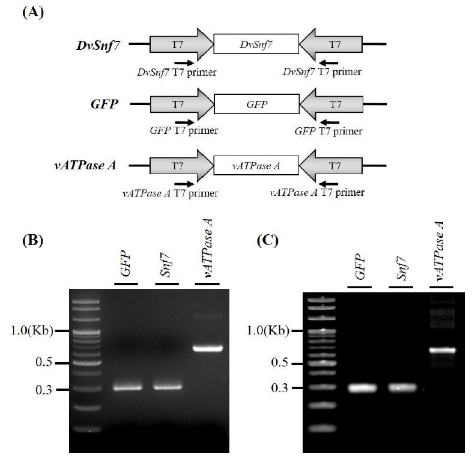
Synthesis of Snf7, GFP, and vATPase A dsRNA and evaluation of their expression in C. septempunctata
Snf7 dsRNA purity is important for conducting toxicity risk assessment for C. septempunctata, as it is not possible to determine the risks posed by insufficiently pure dsRNA. To confirm the purity and identity of the Snf7 dsRNA, the dsRNAs were cloned into L4440 expression vectors (Fig. 4A) and Snf7, GFP (negative control), and vATPase A (positive control) dsRNA were synthesized using RNA polymerase. The synthesized dsRNAs were confirmed by gel electrophoresis. Each dsRNA was cloned and identified as a single band (Fig. 4B). We also confirmed that the dsRNAs were purely synthesized (Fig. 4C). These results suggest that the dsRNA synthesis method used herein can efficiently generate highly purified Snf7 dsRNA. In addition, it can be applied to the synthesis of excess dsRNA required for the risk assessment of imported RNAi based LMOs in domestic insect species.
Fig. 4.
Dietary Snf7 dsRNA risk assessment for C. septempunctata
To evaluate the risk of the synthesized Snf7 dsRNA against C. septempunctata, an experiment was first conducted to ascertain whether Snf7 dsRNA administered to the insects was present in vivo. To confirm the presence of Snf7 dsRNA in vivo, we sampled C. septempunctata on day 1, 3, and 5 after daily dietary treatment with Snf7, GFP, and vATPase A dsRNAs. RT PCR was performed to ensure that the dsRNA showed stable levels in the body of C. septempunctata (Table 3). We confirmed that Snf7, GFP, and vATPase A dsRNA migrated into C. septempunctata and were stable throughout the feeding period (Fig. 5). Based on these results, a risk assessment of the pure synthesized Snf7 dsRNA in C. septempunctata was performed by modifying the previously reported RNAi dsRNA ladybug risk assessment method. The first instar larvae of C. septempunctata were treated with 2 μg/L of Snf7, GFP, or vATPase A dsRNA for 7, 14, and 21 days, and the experiment was repeated thrice (Fig. 6A). The test containers were observed at least once daily for insects showing symptoms and dead ones. The movement of the larvae was observed every day, and if there was no movement or response when touched with a micro brush, the insect was considered dead. We identified non adult individuals and calculated their survival rates. The Snf7 dsRNA treated group showed no differences in survival compared to the negative control (Fig. 6B). In addition, comparing the Snf7 dsRNA treated group with the negative control, the Snf7 dsRNA treatment showed no effect on the development time or dry weight of C. septempunctata (Fig. 6C, D). In contrast, in the vATPase A dsRNA treated group (the positive control), almost all the C. septempunctata died, confirming that the Snf7 dsRNA risk assessment experiments were successful.
Fig. 5.
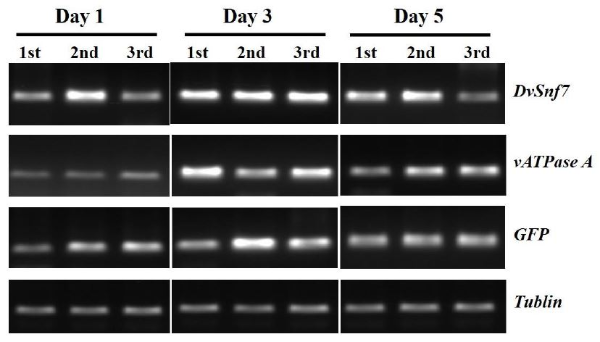
Fig. 6.

The survival rate of C. septempunctata was measured at ea ch growth stage. Each experiment was repeated thrice. (C, D) After treatment with the dsRNAs for 5 days, the developmental time and dry weight of C. septempunctata were measured and compared between the three groups. Each experiment was repeated thrice.
In conclusion, dietary supplementation of Snf7 dsRNA did not pose any risk to C. septempunctata. However, considering the potential domestic environmental and ecological impacts, the risk assessment of C. septempunctata using high concentrations of Snf7 dsRN A requires further research. Therefore, methods for assessing the risks of unintentional spread of LM crops on herbivore predators must be designed and specific RNAi based LM crops must be developed for this purpose. In the future, we plan to develop RNAi based LM crops which will select herbivorous insects eaten by C. septempunctata and develop a food chain based (LM crops herbivorous insects predators) LMO risk assessment method suitable for domestic conditions. The development of risk assessment methods for LMOs for domestic insect species will enable active responses to the risks posed by new LMOs which are rapidly developed and imported to the ecosystem.