Introduction
The genus Medicago L. belongs to the family Fabaceae and consists of approximately 87 species that are predominately distributed from the Mediterranean to Central Asia (Choi et al., 2022; Steele et al., 2010). The genus comprises annual or perennial herbs and a few shrubs, including alfalfa (Medicago sativa) as a widely cultivated forage crop and Medicago truncatula as a legume model plant (Echeverria et al., 2021; Thanopoulos, 2007). Based on studies of flora and distribution, five taxa, including M. sativa, Medicago ruthenica, Medicago polymorpha, Medicago minima, and Medicago lupulina are speculated to be distributed in South Korea (Kil et al., 2004; Kim, 2005; Lee, 1976; Lee et al., 2018; Van Berkum et al., 1998). However, the exact number of taxa present in South Korea remains unknown. These Medicago plants have been considered alien species introduced from Europe, the Mediterranean, and Mongolia to South Korea (Kim, 2005; Lee et al., 2018; Van Berkum et al., 1998). They are usually distributed in open fields, road verges, pastures, parks, and banks (Kil et al., 2004; Kim, 2005; Song & Park, 2019; You, 2018). M. sativa is used as livestock feed, and a living modified (LM) version has been developed recently (Choi et al., 2020; Guertler et al., 2019). This LM organism (LMO) has been approved for use in many countries, including South Korea (Choi et al., 2020). However, concerns regarding potential adverse effects on biodiversity related to alien species and LMOs have been increasing (Kim et al., 2020; Terrones et al., 2021). For the management of alien species or LMOs, rapid and accurate identification should be a priority to prevent adverse effects on biodiversity (Kim et al., 2020; Terrones et al., 2021).
Morphological characteristics such as plant type (herb/shrub), legume type (spiral/not spiral), leaflet or petiole shape, flower color, and number of flowers in each inflorescence are generally evaluated when identifying Medicago species (Chen et al., 2021; Wu et al., 2010). The five taxa of Medicago in South Korea are also identified based on morphological characteristics. M. sativa is a shrub that can be easily distinguished from other species (Wu et al., 2010). M. ruthenica, M. polymorpha, M. minima, and M. lupulina are herbs and they are distinguished based on morphological characteristics such as leaflet or legume shape, and flower color (Wu et al., 2010). However, it is difficult to accurately distinguish the five taxa based on these morphological characteristics outside of their flowering and ripening periods.
Various DNA markers such as random amplified polymorphic DNA, amplified fragment length polymorphisms, single nucleotide polymorphisms, microsatellites, and short sequence repeats are mainly used for studying plant ecology, taxonomy, phylogeny, and genetics (Azizi et al., 2021; Dar et al., 2019; Marakli, 2018). The use of DNA-based markers is advantageous for identifying plant species with similar morphologies as these methods use small amounts of sample material in a time-saving and cost-effective manner (An et al., 2019). Recently, highly variable sequences in plastid genomes have been identified that can facilitate specific plant species identification (Dong et al., 2012; Gao et al., 2011). Analyzing nucleotide sequence data of plastids represents an effective strategy for species identification in plants (Fazekas et al., 2008; Kress & Erickson, 2007).
The target region of the chloroplast genome can be amplified more easily than nuclear DNA when using polymerase chain reaction (PCR) because a plant leaf cell contains up to 10,000 copies of the chloroplast genome (Koya et al., 2005; Lee et al., 2017; Morley & Nielsen, 2016). Although most nucleotide sequences of chloroplast genes are conserved, significant nucleotide variation has been confirmed in chloroplast intergenic spacer regions which allow interspecies comparison (McCauley, 1995). In addition, plastid sequences are generally used to establish molecular markers to identify plants in various genera in ecological and agricultural studies (Abid et al., 2019; Moon et al., 2016; Nguyen et al., 2020; Zhang et al., 2017). However, although species identification is potentially essential for studies on plants, no investigations on species-specific DNA markers in the five aforementioned Medicago plant species have been reported yet.
Taxonomic studies using molecular markers can improve our understanding of the relationship among plant species and can improve management of potentially damaging plant management. In this study, we applied molecular phylogeny based on the nuclear gene gibberellin 3-oxidase 1 (GA3ox1) and chloroplast region tRNALys (UUU) to maturase K (trnK-matK) to five Medicago taxa collected from South Korea. Additionally, we developed a PCR-based species-specific detection method based on plastid DNA sequences to distinguish species with similar morphology.
Materials and Methods
Plant material
Leaves and mature seeds were collected for DNA extraction and PCR analysis. M. polymorpha and M. ruthenica samples were collected from Jindo-gun, Jeollanam-do and from Jeju-si, Jeju-do, respectively. The leaf samples were dried using silica gel and stored at 4°C until DNA extraction. Seeds of M. sativa (NIBRVP0000540343), M. minima (NIBRVP0000499194), and M. lupulina (NIBRVP0000600808) were obtained from the National Institute of Biological Resources (NIBR, Incheon, Korea). Thirty-eight species of Medicago, Melilotus indicus, Melilotus albus, Trigonella elliptica, and Trigonella anguina of Fabaceae were included as outgroups, based on a previous study using trnK-matK and GA3ox1 nucleotide sequence data (Steele et al., 2010). GenBank accession numbers of species used in this study are listed in Appendix 1.
DNA extraction and PCR amplification
Genomic DNA was isolated from fresh tissues using the Libex NP968 system (Tianlong, Xi’an, China) according to the manufacturer’s instructions. PCR amplification was performed using the ProFlex PCR System (Thermo Fisher Scientific Inc., Waltham, MA, USA). PCR reactions were conducted using 2×LAMP Taq PCR Pre-Mix (Biofact, Daejeon, Korea) in 30 µL volumes containing 100 ng of each template DNA and 1 µL of each primer (10 pmoL/µL). For the phylogenetic analysis, PCR conditions and primers for two regions of GA3ox1 and trnK-matK were obtained from a previous study (Steele et al., 2010). PCR condition for species-specific detection method was determined with pre-denaturation at 95°C for 5 minutes, followed by 35 cycles at 95°C for 30 seconds, 59°C for 30 seconds, and 72°C for 1 minutes 30 seconds, with a final extension at 72°C for 7 minutes. PCR products were separated via electrophoresis on 2.0% agarose gels, run in a 1×TAE buffer, and identified using the ChemiDoc XRS+ Imaging System (Bio-Rad, Hercules, CA, USA). The PCR products were sequenced using the amplification primers on an ABI 3730XL system (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
The DNA sequences obtained from the National Center for Biotechnology Information (NCBI) and PCR analysis were aligned using ClustalW and BioEdit version 7.2.5 (Hall, 1999). Phylogenetic tree construction was based on two nucleotide sequences as separated data, the maximum parsimony (MP) method was performed using MEGA 11 (Tamura et al., 2021). The consistency index (CI), retention index (RI), and tree length were calculated using MEGA 11. Five hundred bootstrap replications estimated the internal branch strength of a strict consensus tree to support individual clades (Felsenstein, 1985).
Primer design for detection
A species-specific detection method for the five taxa of Medicago was developed using plastid sequence data obtained from the NCBI (accession numbers: MK460494, MK460497, MK460498, MK460499, and NC053371). Plastid sequences were aligned using the ClustalW multiple alignment program in BioEdit version 7.2.5. The primers were designed using regions showing differences in nucleotide sequence between each species (Table 1).
Results
Distribution and sample information of Medicago
Five species of Medicago distributed in South Korea have been studied previously, however, taxonomic and detailed information remain unavailable (Kil et al., 2004; Kim, 2005; Lee, 1976; Lee et al., 2018; Van Berkum et al., 1998). For performing the taxonomic study of Medicago, distribution-related information was collected as specimen information from the website of Korea National Arboretum and previous studies (Choi et al., 2015; Hwang et al., 2013) (Fig. 1). M. sativa is widely distributed in South Korea, and other species have limited distribution (Fig. 1). In the present study, we obtained three samples of M. sativa, M. minima, and M. lupulina from NIBR (Fig. 1A, D, E). Wild samples of M. polymorpha and M. ruthenica were collected from Jindo-gun, Jeollanam-do and from Jeju-si, Jeju-do, respectively (Fig. 1B, C).
Phylogenetic analysis
To identify the five collected samples, we performed the phylogenetic analysis including the five taxa and other related species of Medicago using nucleotide sequence data of GA3ox1 and trnK-matK (Steele et al., 2010). In total, 44 GenBank-derived sequences of Medicago were used as ingroups for phylogenetic analyses.GenBank data for four taxa ofM. indicus, M. albus, T. elliptica, and T. anguina were used as outgroups (Appendix 1). The GA3ox1 and trnK-matK sequence alignments contained 1,270 and 2,526 characters, 436 (34.3%) and 538 (21.3%) variable sites, and 256 (20.2%) and 256 (10.1%) parsimony informative sites, respectively (Table 1). Phylogenetic analysis of the GA3ox1 and trnK-matK datasets resulted in one equally parsimonious tree (tree length=921 and 770; CI=0.61 and 0.80; RI=0.71 and 0.85, respectively).
The cluster of Medicago was significantly supported (bootstrap value=100) in the phylogenetic trees constructed using GA3ox1 and trnK-matK sequences (Fig. 2). Generally, the same species formed a clade; the relationship between each clade is not clear. In the MP tree using GA3ox1 sequences, except for M. sativa, four different clades of four species were obtained (Fig. 2A). For M. minima and M. lupulina, the collected samples and reference sequences formed clades with 100% bootstrap value, respectively. M. ruthenica and M. polymorpha were supported with 98% and 85% bootstrap values, respectively. The phylogenetic tree based on the trnK-matK region showed higher resolution than that obtained using GA3ox1 (Fig. 2B). M. polymorpha, M. minima, and M. lupulina were separated with 100% bootstrap value. M. sativa and M. ruthenica also formed two clades with 86% and 82% bootstrap values, respectively. In the two trees, the five Medicago species formed distinct clades with over 80% bootstrap value. Therefore, it was confirmed that the five Medicago species were distributed in South Korea. In addition, this indicated that identification of the five taxa using plant material for developing detection methods is highly specific.
Development of a species-specific detection method
Novel primer sets were developed to identify the five Medicago taxa, including M. sativa, M. ruthenica, M. polymorpha, M. minima, and M. lupulina, which are distributed in South Korea (Table 2). All primers were designed using the chloroplast genome of each species to increase specificity (Fazekas et al., 2008). The PCR product sizes were selected over 200 bp for visual analysis on agarose gel. Consequently, five primer pairs were successfully obtained for the five Medicago taxa (Table 2). To increase the primer accuracy for species specificity, they were all designed to be contained within conserved and highly variable regions. The two primer sets, named matK_1 (463 bp) and matK_2 (269 bp), were designed using the matK region and separated the five species into two groups (group 1: M. sativa, M. ruthenica, and M. polymorpha; group 2: M. minima and M. lupulina). The other three primers were also constructed using a non-coding region with highly variable sequences between the two conserved genes. The CP_1 primer set (396 bp) was developed using the nucleotide sequences between ribosomal protein S4 and tRNA-Ser to detect three species (M. sativa, M. ruthenica, and M. minima). The CP_2 (401 bp) and CP_3 (284 bp) primer sets consisted of sequences in the Clp protease proteolytic subunit to tRNA-Asn and NADH dehydrogenase subunit 5 to tRNA-His regions, which can identify M. minima and M. ruthenica, respectively.
To detect multiple primer sets using a single simultaneous PCR assay in the five Medicago taxa, various PCR parameters such as annealing temperature and extension time were tested (data not shown). Consequentially, the primer concentration and PCR condition were determined to increase the specificity of PCR amplification. The agarose gel image showed high efficiency for the developed primer pair as only the expected band was detected for each species (Fig. 3). Further, these results confirmed that all PCR product bands were of their expected size. These results showed that a particular species could be distinguished using a combination of the five designed primer pairs. Therefore, the developed PCR method was considered suitable for qualitative analysis of Medicago species in South Korea.
Discussion
Little is known about the investigation of phylogenetic analysis and species-specific DNA markers in the five Medicago plant species speculated to be distributed in South Korea. Therefore, this study applied molecular phylogeny based on the nuclear gene GA3ox1 and chloroplast region TrnK-matK and developed species-specific markers for five Medicago taxa collected from South Korea. Distribution maps of Medicago plants in South Korea were constructed based on specimen information and previous studies (Fig. 1). In this study, we obtained three seed specimen samples of M. sativa, M. minima, and M. lupulina from NIBR and two wild plant samples of M. polymorpha and M. ruthenica. Previous studies have shown that sequences of trnK-matK and GA3ox1 regions have a phylogenetic resolution at the species level in Medicago (Chen et al., 2021; Hu et al., 2014; Steele et al., 2010). Molecular phylogenetic analyses indicated that the phylogenetic trees constructed using nucleotide sequences (trnK-matK and GA3ox1) showed significant separation between the five taxa of Medicago. The GA3ox1-based tree could not distinguish M. sativa (Fig. 2A). Unlike the GA3ox1-based tree, trnK-matK-based analysis could distinguish all five taxa (Fig. 2B). Previous study results strongly supported our research data (Steele et al., 2010). Nucleotide sequences of the five taxa from previous studies and our collected samples formed clusters for the same species.
The five Medicago taxa distributed in South Korea could be identified using plastid DNA sequences (trnK-matK), but not nuclear DNA sequences (GA3ox1). The discrimination efficiency of the developed PCR markers for the five species can be increased based on differences in plastid DNA sequences. Plastid genomes have been used to develop species-specific markers in various fields, such as identifying species in the family Orchidaceae, genera Pinus and Dendrobium, and invasive aquatic plants (Asahina et al., 2010; Li et al., 2021; Scriver et al., 2015; Wachowiak et al., 2004). As expected, bands of the developed PCR markers in the agarose gel image also showed a clear distinction between the five Medicago species (Fig. 3). This method will facilitate similar future studies on distinguishing domestic and foreign species of Medicago.
In conclusion, we clarified that five species of Medicago are distributed in Korea based on molecular phylogenetic analysis. Our novel PCR-based detection method is suitable for distinguishing Medicago taxa with similar morphologies distributed in South Korea and can be applied to other foreign Medicago species in a timely and cost-effective manner.
Author Contributions
IRK and WC contributed to the conception and design of the study. All authors performed the experiments. IRK, A-MY, and WC analyzed the data. HSL and SL prepared figures and tables. IRK and WC authored or reviewed drafts of the paper. IRK, JRL, and WC participated in valuable discussions. All the authors read and approved the final draft.
References
(1976) Vascular plants and their uses in Korea Bulletin of the Kwanak Arboretum, 1, 1-137 https://s-space.snu.ac.kr/handle/10371/67084.
Figures and Tables
Fig. 1
Maps of Medicago distribution in South Korea. Each map shows the distribution areas (colored patches) and sample collection sites (red circles) of (A) Medicago sativa, (B) Medicago polymorpha, (C) Medicago ruthenica, (D) Medicago minima, and (E) Medicago lupulina.
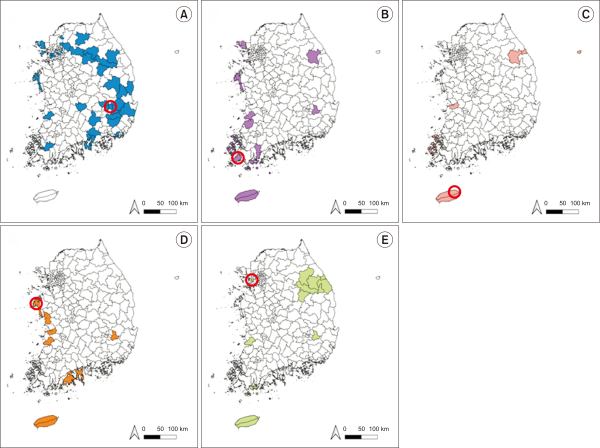
Fig. 2
Molecular phylogenetic tree of Medicago. (A) Maximum parsimony strict consensus tree (tree length=921, CI=0.61, RI=0.71) inferred from GA3ox1 sequence data. (B) Maximum parsimony strict consensus tree (tree length=770, CI=0.80, RI=0.85) inferred from trnK-matK sequence data. Numbers in the branches indicate support values. Red-labelled text indicates samples collected in this study. CI, consistency index; RI, retention index.

Fig. 3
Specificity test of the polymerase chain reaction primers designed for Medicago species in South Korea. Agarose gel images correspond to (A) Medicago sativa, (B) Medicago polymorpha, (C) Medicago ruthenica, (D) Medicago minima, and (E) Medicago lupulina, respectively. Lane M: 100 bp DNA ladder marker; Lane 1: matK_1 primer; Lane 2: matK_2 primer; Lane 3: CP_1 primer; Lane 4: CP_2 primer; Lane 5: CP_3 primer.
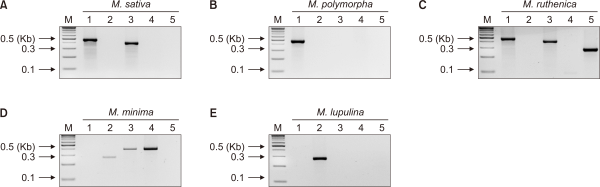
Table 1
Tree statistics of the GA3ox1 and TrnK-matK region based on maximum parsimony analysis
| Characteristics | GA3ox1 | trnK-matK |
|---|---|---|
| Number of OTUs (ingroup/outgroup) |
53 (49/4) | 53 (49/4) |
| Aligned length (bp) | 1,270 | 2,526 |
| Variable characters (%) | 436 (34.3) | 538 (21.3) |
| Parsimony informative characters (%) |
256 (20.2) | 256 (10.1) |
| Length of maximum parsimonious trees |
921 | 770 |
| Consistency index | 0.61 | 0.80 |
| Retention index | 0.71 | 0.85 |
Table 2
List of designed primers and results to identify the five species
| Primer name | Primer sequence (5´–3´) |
Amplification results | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Product size (bp) |
Medicago sativa |
Medicago polymorpha | Medicago ruthenica | Medicago minima | Medicago lupulina | ||
| matK_1_F | CCAAAAACTCGATTTCTATTTTTTCAAAA | 463 | + | + | + | ||
| matK_1_R | TGACTCCGTACCACTGAACG | ||||||
| matK_2_F | TGGGCCGATTCATCCGATTT | 269 | + | + | |||
| matK_2_R | TGAATTGCATTCATTATTCCAATAATTGG | ||||||
| CP_1_F | TACAGCGAAGGGTGAACAAA | 396 | + | + | + | ||
| CP_1_R | CTGATCTCAATGTTTGCAAATAG | ||||||
| CP_2_F | CATTGAAGCGGCTAATCCTATGC | 401 | + | ||||
| CP_2_R | GGCTGACTATACTGCTTTTGGGC | ||||||
| CP_3_F | GTTGTTTATGAATGTCTCATCTGAA | 284 | + | ||||
| CP_3_R | TAGACATAGTATAGGGCGGATGTAG | ||||||