Introduction
The Argentine ant, Linepithema humile, is native to South America but commonly occurs in a Mediterranean climate, mainly in temperate and subtropical regions (Orr & Seike, 1998). It is a polygynous species, with dozens to hundreds of queen ants gathering in the colony and worker ants are brown and uniform in size (Suarez et al., 2001). This species can inhabit various environments, such as piles of stones, under trees, construction materials, and old trees (Hartley & Lester, 2003). They are recognized as one of the world 100 worst invasive alien species by the International Union for Conservation of Nature (2020), and as a dangerous species in Europe, New Zealand, and the United States (Wetterer et al., 2009). Cargo transportation among countries has been reported as the main cause of the invasion by this species, with records of their dissemination by ship to locations such as Hawaii and New Orleans (Suarez et al., 2001). Although damage to humans has not been reported, there are reports of damage to the ants and arthropods living in invaded areas because of the great fertility and adaptability of L. humile (Bolger et al., 2000). The species negatively impacts biodiversity by damaging the seeds and fruits of plants and by having a symbiotic relationship with aphids and scale insects that damage plants, ingesting their secretions while protecting them from their natural enemies (Ness & Bronstein, 2004; Newell & Barber, 1913). In areas where L. humile has invaded and settled, the species competes with native ants and effectively drives them out because of its high adaptability and population density (Cammell et al., 1996; Carpintero et al., 2005; Cole et al., 1992; Ward, 1987). Further, it has also been reported, along with other species, to have actively disseminated Phytophthora citricola, a pathogen responsible for stem cankers in avocado groves in California, USA (El-Hamalawi & Menge, 1996). Additionally, this species act as hosts for various pathogens that cause diseases in bees (Gruber et al., 2017).
L. humile was first discovered in Korea in October 2019, during the inspection of red imported fire ants, and was identified as an ecosystem disturbance organism after undergoing an ecosystem risk assessment by the Ministry of Environment and the National Institute of Ecology (Ministry of Environment, 2020). To date, research on L. humile in Korea includes the first discovery in South Korea reported by Lee et al. (2020). Subsequently its molecular characteristics were studied by Park et al. (2021). However, detailed studies on the habitats and bionomics of this species in South Korea are lacking. Therefore, in this study, we identified the distribution status, host plants, symbiotic species, habitat status, bait preference, and cytochrome c oxidase subunit I (COI) sequence of L. humile collected near the Busan Station in Busan. The results of this study shall be used as a basis to minimize the damage to the Korean ecosystem caused by this invasive species.
Materials and Methods
Species under study
Workers, males, and queens L. humile ants were collected and bred, with the permission of the commission of the competent local environmental agency, in accordance with Article 24 of the 「Act on the Conservation and Use of Biological Diversity」 (Fig. 1).
Survey area and specimen collection
To confirm the habitat of L. humile, an area with a 2-km radius near Busan Station (35° 06’ 54’’, 129° 02’ 32’’) was surveyed each month from October 2019 to April 2021, focusing on areas where ants formed colonies near roads, parking lots, residential areas, parks, and container yards (Fig. 2), through visual inspection and using pitfall traps. For visual inspection, tweezers and an insect pooter were used at locations where ant nests formed small heaps of dirt. For inspection using pitfall traps, a total of 900 traps were installed monthly (50 per month) at 30-50 m from each other; meat bait (cat food) was placed in 50-mL conical tubes in which a 2-mm diameter hole was drilled (Fig. 3). An attractant prepared with eco-friendly antifreeze and ethanol (ethyl alcohol 95%) mixed at a 1:1 ratio was used to use to kill and preserve the specimens. The global positioning system coordinates for the survey area were marked using satellite images from ArcGIS version 10.5 ( https://www.arcgis.com/index.html). Collected specimens were classified and identified (SZX16; OLYMPUS, Tokyo, Japan) in the laboratory of the National Institute of Ecology (Room No. 219) before being photographed (DP27; OLYMPUS) and then stored in a liquid medium.
Surveying the distribution area of L. humile in the vicinity of the site of its discovery
Flora and host plants
From October 2020 to July 2021, plants distributed along roads, flower gardens, and residences in the vicinity of the habitat of L. humile were surveyed, and a list of plants that served as food was compiled. The illustrated books by Lee (2003) and Park (2009) were used to identify the plants. For scientific names and classification system, the National Institute of Biological Resources (2021) was consulted. Alien and invasive species classification followed the National Institute of Ecology (2021), and planted species such as roadside trees were indicated separately.
Symbiotic species
Aphids and symbiont host plants found in the habitat of L. humile were photographed using a digital camera (Eos 5D Mark III; Canon, Tokyo, Japan), collected, identified, and stored in a liquid medium. Aphid identification was based on the illustrated book of Korean aphids from Seo et al. (2019).
Competition with native species
The collected L. humile specimens were identified using a stereomicroscope (SZX16; OLYMPUS) and then dried (exsiccata). The species dominance in each trap was calculated using ggplot Ribbon in R studio version 4.1.2 ( https://cran.r-project.org/bin/windows/base/old/4.1.2/), as follows:
Bait preference test
Queen and worker ants were separated into four colonies in transparent polycarbonate containers (32.5× 26.5×15 cm) for the experiment, which was repeated four times. Emu oil was applied at the entrance of the polycarbonate container as a natural escape-prevention agent. Only water was provided until the experiment was started (Enzmann et al., 2012), and the methods commonly used in ant exploitation and interference competition were used as references (Krushelnycky & Reimer, 1998). Ten types of bait (egg yolk, egg white, yogurt+glucose, egg yolk+glucose+yogurt, water+glucose [25%], jelly, grain cracker crumbs, yogurt, tuna feed, and minced mealworm) were used in the experiment. To confirm the preferred bait proportions, L. humile individuals were observed at 10-minute intervals, six times per experiment, with four repetitions.
Mitochondrial cytochrome c oxidase subunit-I sequence analysis
Twenty specimens collected from the area around Busan Station were stored in 95% ethanol. Genomic DNA was extracted using MG Tissue Genomic DNA Extraction SV (MD014; Doctor Protein Inc., Seoul, Korea). The mitochondrial COI sequence was amplified using Dr. MAX DNA Polymerase (DR00302; Doctor Protein Inc.) and the primers LCO1490-JJ (5ʹ-CHACWAAYCATAAAGATATYGG-3ʹ) and HCO2198-JJ (5ʹ-AWACTTCVGGRTGVCCAAARAATCA-3ʹ; Astrin & Stüben, 2008). The PCR reaction system was set as follows: denaturation at 95°C for 5 minutes, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 45°C, and 1 minute at 72°C for extension, and a final extension at 72°C for 7 minutes. The BigDye® Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA) was used for sequencing, and the species was identified using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) database. The obtained sequence was compared with 29 sequences of L. humile samples from the NCBI database in a neighbor-joining analysis using the Tamura-Nei model with 1,000 bootstraps (MEGA 7.0; Kumar et al., 2016). Linepithema micans (430 bp) was used as an out-group.
Results
Current distribution of L. humile in Korea
The distribution of L. humile was confirmed at approximately 1.5 km along the road near Busan Station. Colonies were found around stone crevices, railroads, underpasses, and containers, and also in areas in which human activities were present, such as inside Busan Station, parking lots, and buildings (Fig. 4). The habitat of Argentine ants tended to spread around September 2020 but in 2021 there were places that were fragmented due to road construction and street tree movement in the Busan Station area.
Current status of the area in the vicinity of the point of discovery of L. humile
Flora and feeding activity
A total of 63 species from 37 families were identified among the flora from the area inhabited by L. humile, and nine species from seven families were confirmed as host plants: Japanese ivy (Hedera rhombea), paperplant (Fatsia japonica), Indian chrysanthemum (Dendranthema indicum), hardy garden mum (Chrysanthemum morifolium), lily turf (Liriope platyphylla), glossy privet (Ligustrum lucidum), lawn grass (Zoysia japonica), camellia (Camellia japonica), and zelkova (Zelkova serrata) (Table 1). A total of 35, 28, and 37 herbaceous, woody, and planted species were found, respectively. Feeding activity was verified in four herbaceous and five woody species. No preference of the ants for specific plant families was observed, and they were typically concentrated in herbaceous or woody plants with flowers or flower buds. Fourteen alien plants and two invasive species (Ecosystem-disturbing species) were identified among the 63 species recorded.
Symbiotic species
Four species of aphids were identified in the areas inhabited by L. humile: cotton aphid (Aphis gossypii) and mango aphid (Aphis odinae) on Indian chrysanthemum (Dendranthema indicum) and Japanese euonymus (Euonymus japonicus), black bean aphid (Aphis fabae) on paperplant (Fatsia japonica), and brown citrus aphid (Toxoptera aurantii) on camellia (Camellia japonica) (Table 2, Fig. 5).
Status of ant habitats
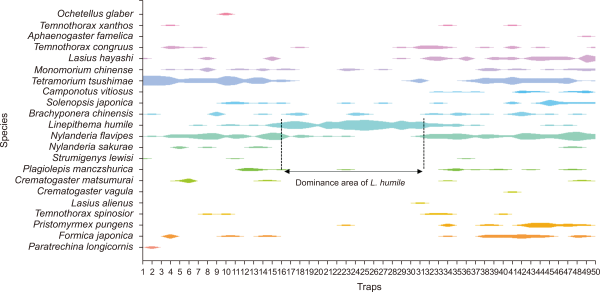
A total of 22 species within 17 genera from 4 subfamilies were identified in the survey. Linepithema humile was the most frequently found species in the survey with traps, followed by the Korean native species Tetramorium tsushimae and Nylanderia flavipes. In traps where L. humile was dominant, some Korean species including N. flavipes, T. tsushimae, and Monomorium chinense were identified, and the highest proportion of T. tsushimae was found in non-dominant areas. L. humile succeeded in the competition with Korean native species, and a tendency to decrease by one third was observed in the populations of the latter. The reduction of species diversity of Korean ants was confirmed by the analysis using R studio version 4.1.2 (Fig. 6).
Bait preference test
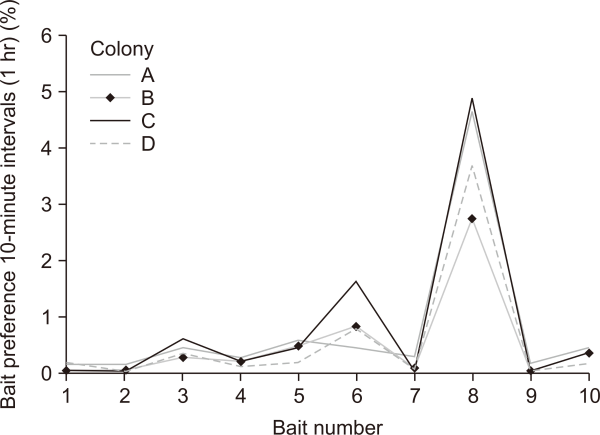
In the experiment to determine bait preference of worker and queen ants, only the former consumed the bait in most tests, while some fed through trophallaxis. After six sessions at 10-minute intervals, bait preference recorded was in the order: jelly>water+glucose (25%)>yogurt. Most of the preferred bait had high sugar content, and all four colonies showed the highest preference for jelly. Tuna feed, which is rich in protein but is not easy to consume rapidly due to its solid state, had the lowest preference rates (Table 3, Fig. 7).
COI sequence analysis
All 630-bp COI sequences from the 20 L. humile individuals from different occurrence points were identical, confirming that a single population disseminated after invasion into Korea. The BLAST results of the 630-bp COI sequence were 99% identical to L. humile (Table 4).
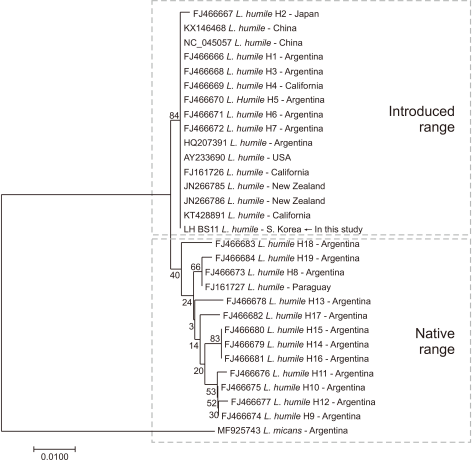
The neighbor-joining analysis showed that L. humile from Korea shared the same sequence with individuals in non-native invaded areas, such as China, Japan, and New Zealand. Furthermore, these individuals were separated into groups according to their origin (Fig. 8).
Discussion
L. humile has been reported as having dispersed from several to several thousand kilometers by human transport over the last 100 years, with records of dissemination by ship transport from invaded areas such as Hawaii and New Orleans (Carpintero et al., 2005; Harris, 2002). As the species does not perform nuptial flights, its mobility is low and, consequently, its worldwide spread occurs indirectly in association with movement owing to trade among countries (Sanders et al., 2001). Indeed, it is more than likely that L. humile was unintentionally introduced in Korea by shipping containers. As a highly adaptable and invasive species, it settles in urban and suburban areas with low vegetation density and succeeds at exploitation and competition by expelling native ants (Holway, 1999; Human & Gordon, 1996). Therefore, an ecosystem imbalance in Korea is expected.
L. humile prefers temperate climate zones with high humidity, and the optimum spawning temperature is 28°C; thus, it is most active during spring and autumn (Abril et al., 2008). In 2016-2021, the average monthly maximum temperatures and average monthly minimum temperatures in Busan were approximately 28°C and 22°C in summer, respectively (AKorea Meteorological Administration, 2020), offering suitable conditions for spawning and feeding activities of L. humile.
The COI sequences from L. humile individuals collected in the Busan Station area were 99% identical to those in the NCBI database. The Korean populations were found to be genetically close to individuals from invaded areas such as China, Japan, California, and New Zealand. Previous studies have reported the absence of attacks among newly introduced L. humile individuals that do not inhabit the same habitat (Giraud et al., 2002). Therefore, the genetic similarity among individuals introduced to other areas is probably high.
A total of nine species within families of plants fed by L. humile were identified, and among them, Chrysanthemum morifolium, Fatsia japonica, and Camellia japonica had a symbiotic relationship with four species of aphids. Damage to crops has been reported because L. humile ingests secretions while protecting aphids and scale insects, which are agricultural pests (Carroll & Dale, 2021). Moreover, the increasing number of aphids causes damage to host plants (Ness & Bronstein, 2004; Newell & Barber, 1913). Further, in South Africa, native ants that aid in the reproduction of the shrub plant Fynbos have reportedly been displaced, and the migration of seeds of Dendromecon rigida has been reduced, compared to regions inhabited by Pogonomyrmex subnitidus (Carney et al., 2003).
L. humile has also invaded Doñana National Park in Spain and occupied 115 of 182 cork trees (Quercus suber), reducing the population of the native Camponotus scutelaris (Carpintero et al., 2005). Therefore, it is only reasonable to expect that, if the population of L. humile increases, damage will be caused to Korean food plants and crops by protecting aphids, causing an imbalance in the ecosystem by reducing native ants related to seed dispersion.
There are several reports of L. humile competing with most of the native ant species, driving them out with its high adaptability and population density, rapidly dominating the areas it invades (Carpintero et al., 2005). Moreover, the displacement of Tetramorium tsushimae, Nylanderia flavipes, and Lasius hayashi from areas in Korea by L. humile has been reported. Recycled waste, wood planks, and piles of factory materials are commonly moved from the areas inhabited by L. humile to treatment plants throughout the country, and trees planted in the soil of these areas are moved or transplanted to other areas. As reported by Sanders et al. (2001), the dissemination of L. humile is highly likely due to unintentional moving by humans, and such spread of this species may lead to a future imbalance in the native ecosystem.
L. humile is highly reproductive and adaptable. Therefore, unless there is proper control after initial detection, effective control will become increasingly difficult, as reported in Japan and California (Xu et al., 2022). Therefore, continuous monitoring of areas suitable for habitat, such as ports and container logistics, is necessary, including areas already invaded by L. humile. Furthermore, this challenge demands a concerted response by all related organizations and institutions.
Figures and Tables
Fig. 1
Adult specimens of Linepithema humile. (A) worker, (B) male, and (C) queen. Scale bar: 1.00 mm.

Fig. 3
Visual inspection and inspection using pitfall traps. (A) Visual inspection using pooters, (B) inspection using soil pitfall traps, (C) inspection using pitfall traps placed in a wire mesh, and (D) diagram of the trap.

Fig. 4
Distribution of Linepithema humile. (A) January 2020, (B) March 2020, (C) September 2020, (D) March 2021, (E) June 2021, and (F) September 2021.

Fig. 5
Aphids present in areas inhabited by Linepithema humile. (A) Aphis gossypii, (B) Aphis fabae, (C) Aphis gossypii and Aphis odinae, and (D) Toxoptera aurantii.
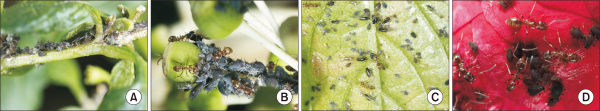
Table 1
Flora of the area inhabited by Linepithema humile
| Family | Scientific name | Life-form | Planted species | Alien species |
Ecosystem-disturbing species | Host plant |
|---|---|---|---|---|---|---|
| Aizoaceae | Lampranthus spectabilis (Haw.) N.E.Br. | Herbaceous | ○ | |||
| Amaranthaceae | Amaranthus viridis L. | Herbaceous | ○ | |||
| Amaryllidaceae | Narcissus tazetta subsp. chinensis (M.Roem.) Masam. & Yanagita | Herbaceous | ○ | |||
| Apocynaceae | Vinca major L. | Woody | ○ | |||
| Aquifoliaceae | Ilex crenata Thunb. | Woody | ○ | |||
| Araliaceae | Fatsia japonica (Thunb.) Decne. & Planch. | Woody | ○ | ○ | ||
| Hedera rhombea (Miq.) Siebold & Zucc. ex Bean | Woody | ○ | ○ | |||
| Arecaceae | Trachycarpus fortunei (Hook.) H.Wendl. | Woody | ○ | |||
| Asteraceae | Artemisia indica Willd. | Herbaceous | ||||
| Chrysanthemum morifolium Ramat. | Herbaceous | ○ | ○ | |||
| Conyza canadensis (L.) Cronquist | Herbaceous | ○ | ||||
| Conyza sumatrensis (Retz.) E. Walker | Herbaceous | ○ | ||||
| Coreopsis lanceolata L. | Herbaceous | ○ | ||||
| Dendranthema indicum (L.) Des Moul. | Herbaceous | ○ | ||||
| Farfugium japonicum (L.) Kitam. | Herbaceous | ○ | ||||
| Lactuca scariola L. | Herbaceous | ○ | ○ | |||
| Senecio vulgaris L. | Herbaceous | ○ | ||||
| Solidago altissima L. | Herbaceous | ○ | ○ | |||
| Sonchus oleraceus L. | Herbaceous | ○ | ||||
| Taraxacum officinale F.H.Wigg. | Herbaceous | ○ | ||||
| Youngia japonica (L.) DC. | Herbaceous | |||||
| Aucubaceae | Aucuba japonica f. variegata (Dombrain) Rehder | Woody | ○ | |||
| Berberidaceae | Nandina domestica Thunb. | Woody | ○ | |||
| Buxaceae | Buxus microphyulla var. koreana Nakai ex Rehder | Woody | ○ | |||
| Cannaceae | Canna × generalis L.H. Bailey | Herbaceous | ○ | |||
| Caprifoliaceae | Abelia × grandiflora (Rovelli ex André) Rehder | Woody | ○ | |||
| Viburnum odoratissimum var. awabuki (K. Koch) Zabel ex Rümpler | Woody | ○ | ||||
| Caryophyllaceae | Stellaria media (L.) Vill. | Herbaceous | ||||
| Sagina japonica (Sw.) Ohwi | Herbaceous | |||||
| Celastraceae | Euonymus japonicus Thunb. | Herbaceous | ○ | |||
| Euonymus alatus (Thunb.) Siebold | Woody | ○ | ||||
| Convolvulaceae | Ipomoea nil (L.) Roth | Herbaceous | ○ | |||
| Cupressaceae | Juniperus chinensis ‘Kaizuka’ | Woody | ○ | |||
| Ericaceae | Rhododendron indicum (L.) Sweet | Woody | ○ | |||
| Lamiaceae | Lamium amplexicaule L. | Herbaceous | ||||
| Liliaceae | Liriope platyphylla F.T. Wang & T. Tang | Herbaceous | ○ | ○ | ||
| Yucca gloriosa L. | Woody | ○ | ||||
| Lythraceae | Lagerstroemia indica L. | Woody | ○ | |||
| Malvaceae | Althaea rosea (L.) Cav. | Herbaceous | ○ | |||
| Oleaceae | Ligustrum lucidum W.T. Aiton | Woody | ○ | ○ | ||
| Ligustrum obtusifolium Siebold & Zucc. | Woody | ○ | ||||
| Osmanthus heterophyllus (G. Don) P.S. Green | Woody | ○ | ||||
| Onagraceae | Oenothera biennis L. | Herbaceous | ○ | |||
| Oxalidaceae | Oxalis corniculata L. | Herbaceous | ||||
| Pinaceae | Pinus thunbergii Parl. | Woody | ○ | |||
| Pittosporaceae | Pittosporum tobira (Thunb.) W.T. Aiton | Woody | ○ | |||
| Poaceae | Bromus catharticus Vahl | Herbaceous | ○ | |||
| Elymus tsukushiensis Honda | Herbaceous | |||||
| Festuca arundinacea Schreb. | Herbaceous | ○ | ||||
| Poa annua L. | Herbaceous | |||||
| Sasa borealis (Hack.) Makino & Shibata | Herbaceous | ○ | ||||
| Setaria viridis (L.) P. Beauv. | Herbaceous | |||||
| Zoysia japonica Steud. | Herbaceous | ○ | ○ | |||
| Polygonaceae | Rumex crispus L. | Herbaceous | ○ | |||
| Rosaceae | Photinia glabra (Thunb.) Maxim. | Woody | ○ | |||
| Rubiaceae | Galium spurium L. | Herbaceous | ||||
| Gardenia jasminoides J. Ellis | Woody | ○ | ||||
| Sapindaceae | Koelreuteria paniculata Laxm. | Woody | ○ | |||
| Solanaceae | Solanum nigrum L. | Herbaceous | ||||
| Taxaceae | Taxus cuspidata Siebold & Zucc. | Woody | ○ | |||
| Theaceae | Camellia japonica L. | Woody | ○ | ○ | ||
| Ulmaceae | Zelkova serrata (Thunb.) Makino | Woody | ○ | ○ | ||
| Vitaceae | Parthenocissus tricuspidata (Siebold & Zucc.) Planch. | Woody | ○ |
Table 2
Symbiotic relationship of aphids with Linepithema humile
| Family | Scientific name | Common name | Feeding activity |
|---|---|---|---|
| Aphididae | Aphis gossypii | Cotton aphid | Chrysanthemum (Dendranthema indicum) |
| Aphis odinae | Mango aphid | Japanese euonymus (Euonymus japonicus) | |
| Aphis fabae | Black bean aphid | Paperplant (Fatsia japonica) | |
| Toxoptera aurantii | Brown citrus aphid | Camellia (Camellia japonica) |
Table 3
Bait preference of Argentine ants (Linepithema humile) at 10-minute intervals for one hour
| Bait number | Colony (queen/worker) (%) | ||||
|---|---|---|---|---|---|
|
|
|||||
| A (20/250) |
B (20/250) |
C (20/250) |
D (20/250) |
E (20/250) |
|
| 1. Egg yolk (liquid) | 0.17 | 0.22 | 0.05 | 0.05 | 1.58 |
| 2. Egg white (liquid) | 0.13 | 0.02 | 0.03 | 0.03 | 0.17 |
| 3. Yogurt+glucose (liquid) | 0.47 | 0.38 | 0.62 | 0.62 | 0.25 |
| 4. Egg yolk+glucose+yogurt (liquid) | 0.30 | 0.12 | 0.23 | 0.23 | 0.42 |
| 5. Yogurt (liquid) | 0.58 | 0.20 | 0.47 | 0.47 | 0.58 |
| 6. Water+glucose (25%, liquid) | 0.47 | 0.78 | 1.63 | 1.63 | 0.79 |
| 7. Minced mealworm (solid+liquid) | 0.32 | 0.05 | 0.05 | 0.05 | 0.58 |
| 8. Jelly (semisolid) | 4.68 | 3.70 | 4.90 | 4.90 | 3.92 |
| 9. Tuna feed (solid) | 0.18 | 0.03 | 0.03 | 0.03 | 0.79 |
| 10. Grain cracker crumb (solid) | 0.47 | 0.18 | 0.38 | 0.38 | 1.88 |
Table 4
NCBI-based BLAST results of the COI sequence of Linepithema humile
| Scientific name | Query coverage (%) | Identity (%) |
Accession number |
|---|---|---|---|
| Linepithema humile |
100 | 100.00 | MT890564 |
| Linepithema humile |
100 | 99.84 | NC_045057 |
| Linepithema humile |
100 | 99.84 | MT606344 |
| Linepithema humile |
100 | 99.84 | JN266786 |
| Linepithema humile |
100 | 99.84 | JN266785 |
| Linepithema humile |
100 | 99.84 | HQ207391 |