Introduction
Limax maximus (Gastropoda: Limacidae) is recognized as one of the world’s most aggressively invasive terrestrial mollusks (Barker & McGhie, 1984; Barker, 1999). Originating from Europe, North Africa, and Asia, L. maximus has become an invasive species. It has successfully colonized various countries and islands, including, but not limited to, North America, South America, Africa, the Pacific Islands, Australia, Hawaii, New Zealand, and Japan (Barker & McGhie, 1984; Roth & Sadeghian, 2006; Joe & Daehler, 2008; Hasegawa et al., 2009). L. maximus exhibits omnivorous tendencies, with a diet comprising dead plant matter and fungi. It is considered to have an impact on plant diversity through selective feeding (Buschmann et al., 2005; Strauss et al., 2009). Notably, this species is regarded as a major agricultural pest in Florida, USA, particularly on cucumber, sweet potato, lettuce, Chinese cabbage, and horticulture plants, likely favoring tender, leafy vegetables, and other soft-stemmed plants (Stange et al., 2009). Furthermore, in regions such as Hawaii and New Zealand, L. maximus and other slugs have been reported to be direct causes of local plant extinctions (Joe & Daehler, 2008).
Two other slug species in the genus Limax, namely L. flavus and L. marginatus, have been unintentionally introduced to the Republic of Korea (Lee et al., 2010). However, to date, there is no documented evidence of established L. maximus populations in the Republic of Korea (CABI Digital Library, n.d.; GBIF Secretariat, 2023; National Institute of Biological Resources (NIBR), n.d.). The first documented occurrence of L. maximus in Japan was reported in 2006 (Hasegawa et al., 2009). Since then, it has spread to Hokkaido and mainland Japan (Morii & Nakano, 2017). This implies that L. maximus could potentially thrive in all regions of Republic of Korea, given that Japan and Korea have similar latitude and environmental conditions.
This study aimed to provide the first confirmed reports of L. maximus in Republic of Korea and furnish ecological and molecular data that could aid in the effective management of indigenous natural ecosystems following its introduction. Specifically, this research focused on barcoding analysis for the identification and classification of L. maximus, offering fundamental information necessary for understanding its potential impact within ecosystems and for the development of management strategies.
Materials and Methods
Site description
Suwon-si is a special case city covering an area of 121 km² in the inland region of southwestern Gyeonggi-do, Republic of Korea. It is characterized by a gentle slope running from northeast to southwest. The topography to the south predominantly consists of plains, lacking high or rugged mountains in the vicinity. The area experiences a continental climate with an average annual temperature of 12.5°C, hot summers, and relatively cold winters according to data from www.weather.go.kr. The survey was conducted three times daily and nightly between August and September 2023, focusing around Bambat Tree Frog Park in Suwon (N37˚18’25.46”, E126˚58’16.1”), where the initial sighting of L. maximus occurred. The survey area characterized by forested landscapes has hobby farms, agricultural farms, and trails. It is nestled by mountains connecting to Deokseong Mountain (160 m above sea level), with the Bamkkot Village apartment complex within a 100 m radius. This proximity makes it a favored spot for hiking and nature exploration (Fig. 1).
Sample collection and preservation
Slugs were collected alive, one at a time. They were immediately submerged in 70% ethanol and transported to the laboratory. Upon arrival, tissues were carefully excised using sterilized surgical scalpels for further analysis.
DNA sequence analysis
A 50 mg tissue specimen was excised from a L. maximum slug sample using a sterilized knife and transferred into a 3 mm ceramic bead tube. To enhance DNA extraction efficiency from the slug sample, a modification was made to the Clear-S™ Quick DNA extraction kit’s manual (IVT3002, Invirustech, Republic of Korea), which was then used for the extraction process. In short, 210 µL of HBS buffer and 10 µL of TCEP-HCl were added to the tube containing the 50 mg sample. Using a homogenizer (MB-24YS, Biobase, China), the sample was repeatedly crushed three times at 50 Hz for 30 s. To the crushed sample, 20 µL of Proteinase K (20 mg/mL) was added. The mixture was then incubated at 55°C for 30 min. Following this, 490 µL of DLD buffer was added. After thorough mixing, the mixture was loaded into a spin column. Subsequent steps were executed in accordance with the kit’s manual. DNA extraction results were assessed based on absorbance, with the OD260/280 value measuring 2.075. The yield was determined to be 0.55 µg.
Following DNA extraction, a polymerase chain reaction (PCR) was carried out to amplify the mitochondrial COI region with a length of approximately 1340 bp of the slug. The PCR process consisted of two stages. In the initial stage, primers mtCOI-1F-54 (5’-tttcaacaaaycataargatatt gg-3’) and mtCOI-1R-53 (5’-aayaccaatagaaattatag cataaa-3’) were used to amplify the first fragment. The second stage used mtCOI-2F (5’-ttagcrggggcaattactatrc-3’) and mtCOI-2R (5’-cgaaaaca-gatattaacgaaccat-3’) to amplify the second fragment. Each primer was designed with reference to the slug COI universal primer developed by Nitz et al. (2009). EzPCR™ Basic 5X Master mix was used to carry out the PCR process, starting with an initial denaturation step at 92°C for 4 min, followed by 40 cycles of amplification at 92°C for 1 min, 50°C for 1 min, and 72°C for 1 min. Following the completion of the PCR cycling, a final extension step was carried out at 72°C for 5 min. Then 2 µL of amplicon was mixed with 0.5X TBE buffer and subjected to 1.5% agarose gel electrophoresis at 150 V for 30 min. A Clear-S™ PCR/Gel DNA fragment purifycation kit (IVT3005, Invirustech, Republic of Korea) was used for sequence confirmation and purification of the PCR product. The purified DNA was then sent to Bionics, Inc. (Daejeon, Republic of Korea) for Sanger sequencing. After obtaining sequences for both the first and second fragments, a consensus sequence was obtained using CAP3, yielding a mtCOI DNA sequence of 1381 bp in length.
Following electrophoresis of PCR amplicons on a 1.5% TBE agarose gel for DNA sequence analysis, lane 1 was loaded with a 100 bp DNA ladder, serving as a reference for molecular size. Lane 2 was loaded with PCR amplicons using the mtCOI-1F-54 and mtCOI-1R-53 primer set 1, and lane 3 was loaded with PCR amplicons using the mtCOI-2F and mtCOI-2R primer set 2. Electrophoresis revealed a distinct band around the target size of 830 bp in lane 2, accompanied by another band near the target size of 800 bp (Fig. 2a). These amplicons were then sequenced and assembled using the CAP3 software. Approximately 1381 bp of the slug mtCOI gene sequence was acquired for phylogenetic analysis, which was aligned using ClustalO. Following alignment, regions with gaps in the bilateral sequence were trimmed, resulting in a final gene sequence of 1231 bp, which was used for subsequent alignment and phylogenetic analysis. Using the program MEGA-X (Kumar et al., 2018), a phylogenetic tree was constructed using the maximum likelihood method based on the Kimura 2-parameter model (Kimura, 1980; Kumar et al., 2018). This process was repeated 100 times to generate a bootstrap consensus tree, clustering taxa based on maximum similarity.
Results and Discussion
Through daily and nightly surveys, a total of 132 slugs were collected from the survey area. They exhibited a tendency to hide in various microhabitats in and around the farm environment, often under awnings and fences during the daytime. Conversely, a large number of individuals were collected along trails leading to nearby fields and near gullies during nocturnal surveys (Table 1). Additionally, these slugs showed increased activity levels during cloudy and rainy weather conditions, resulting in a notable increase in the number of specimens collected during these weather conditions and at night, consistent with their habits elsewhere (Bailey, 1981). In total, 97 (73.5%) slugs were collected along the trail connecting the farmland with the forested region, while 24 (18.2%) slugs were collected from within farms and fences, 15 (11.4%) slugs from the forested area, and two (1.5%) slugs from grassy plains (Table 1). These results indicate that the habitat range of L. maximus in Republic of Korea is quite broad, including gardens, parks, and greenhouses, consistent with a previous report (Weigand, 2014).
L. maximus, which is of the same genus as the domestically recorded species L. flavus and L. marginata in Korea, stands out due to its size and coloration. The largest L. maximus specimen collected during this study measured 150 mm, showcasing no significant size deviation from specimens reported in Japan (Morii & Nakano, 2017). Unlike the domestic species, L. maximus exhibited a larger size, with a body color ranging from yellowish-white to grey and brown. This species can be distinguished by a distinct irregular pattern of brown spots and blotches extending from the head to the tail, including the mantle (Fig. 2b), making it larger than the domestic record species. In comparison, L. flavus, known as medium-sized slugs with an average body length of 130 mm, display a yellowish body adorned with subtle brown spots. Meanwhile, L. marginatus categorized as small slugs with an average body length of 50 mm, feature two distinct black stripes running along their body, distinguishing them from L. maximus (Kwon et al., 2001).
Results revealed that Limax maximus S. Korea (2023), which represented the first discovery of this species in South Korea as identified in this survey, formed a phylogenetic clade with other L. maximus (Nitz et al., 2009) (Fig. 3). In a BLASTn analysis, genes closely matched with those of the L. maximus isolate L1185 COI gene (FJ606469.1), sharing 99.7% sequence similarities and a query cover of 100%. This evidence, along with their appearance, confirmed that these slugs were L. maximus. Originally, L. maximus was primarily found in various forested areas across Europe.
However, its habitats have gradually expanded into anthropogenic environments, including urban and rural gardens, abandoned structures, parks, and greenhouses (Weigand, 2014). In Japan, this species was initially documented in 2012 within a remote natural forest in the Maruyama Forest Park in Sapporo, Hokkaido. Remarkably, within just six years, L. maximus was recorded in an area approximately 100 km from Sapporo (Morii & Nakano, 2017). Since then, this species has spread further into neighboring regions, including the Honshu, Kanto, and Chugoku prefectures (GBIF Secretariat, 2023). Given that Republic of Korea and Japan share similar latitude, it is reasonable to assume that L. maximus has the potential to inhabit the landscape of the Republic of Korea. As L. maximus has been classified as an invasive species in South Africa and Hawaii, USA (CABI Digital Library, n.d.) with potential to prey on and consume native slug species, it is important to consistently monitor the spread of this species (Byers, 2000; Paustian & Barbosa, 2012; Rollo & Wellington, 1979). As highlighted above, distinct characteristics of L. maximus, such as its size and appearance, make it easy to distinguish it from native slug species and other Limax species in Republic of Korea. Thus, the use of citizen science, as has been successfully used in Japan (Morii & Nakano, 2017), could be an effective way to manage the establishment and spread of L. maximus.
Figures and Tables
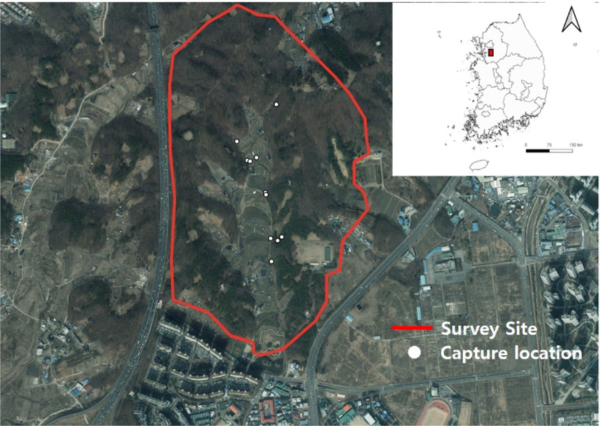
Survey site and capture locations of Limax maximus at the Bambat Tree Frog Park, Suwon-si, Republic of Korea.
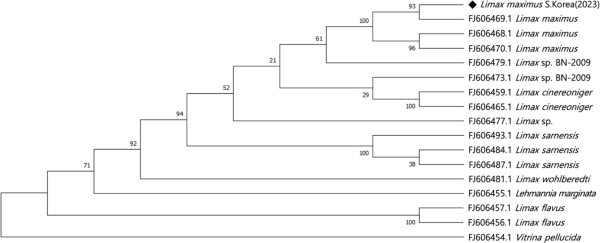
Results of phylogenetic and sequence similarity molecular analysis of Limax maximus. Numbers on nodes indicate bootstrap support values.

