Introduction
Feces is an attractive source of samples for the conservation genetic studies of free-ranging animals because of the ease of sampling and unprecedented sample size without any need to capture or observe the animals (Taberlet and Luikart, 1999; Wehausen et al., 2004). A number of studies have been conducted using these non-invasive samples in a variety of mammalian species, such as primates (baboon, bonobo, langur monkey, and orangutan), carnivores (badger, brown bear, canids, cougar, lynx, mustelids, otter, and seals), herbivores (bats, bighorn sheep, elephant, red deer, rabbit, reindeer, and rhinoceros), sirenia (dugong), cetacea (dolphin), and marsupials (quokka and wombat) in the wild (Waits and Paetkau, 2005).
Many non-invasive sampling studies were aimed at examining various factors affecting DNA extraction yield, such as temperature during collection (Nsubuga et al., 2004), the condition of the samples (Lucchini et al., 2002), and the sampling season (Lucchini et al., 2002), and comparing the efficiency of the sample storage methods (Nsubuga et al., 2004) and DNA extraction protocols (Nsubuga et al., 2004; Wehausen et al., 2004). Also, different fecal pellet materials (outer, inner, and whole pellet) and different amounts of fecal pellet materials have been used to evaluate the rate of DNA amplification success (Wehausen et al., 2004). However, non-invasive studies that quantitatively compared different DNA amplification success of different markers, including studies that compared DNA amplification success between different types of primer set or lengths of the amplified fragments, have rarely been conducted in wildlife populations (Taberlet and Luikart, 1999; Byrne et al., 2021). Addressing these issues seems fundamental but is essential to planning a new research project related to wildlife conservation genetics.
In this review, we selected the long-tailed goral (Naemorhedus caudatus) as a target species for our analyses. The endangered long-tailed goral is a member of the genus Naemorhedus, tribe Rupicaprini, and subfamily Caprinae (Min et al., 2004; An, 2006; Kim, 2021 and 2022). The goral species is registered in the International Union for Conservation of Nature Red List of Threatened Species and Convention on International Trade in Endangered Species Appendices I (Lee et al., 2019) and is distributed across northeast Asia, including Russia, China, and Korea (Lee et al., 2019). Their population size was presumed to be about 800 individuals in South Korea after a massive decline due to illegal hunting, over-exploitation, habitat destruction, and habitat fragmentation (Ministry of Environment 2002; Yang, 2002; Kim, 2021 and 2022). The population size of gorals has increased recently due to policies protecting the endangered species and their habitats, albeit slowly (Lee et al., 2019).
The goal of this review was to provide researchers with efficient guidelines for initiating a new non-invasive study of a variety of wildlife species, particularly endangered species.
Review of Previous Non-Invasive Studies
In this review, we referred to previous studies using non-invasive fecal sampling. In particular, three critical goral papers for mitochondrial DNA (Kim, 2021), microsatellite DNA (Kim, 2022), and sex-linked DNA marker (Kim et al., 2008) were reanalyzed. For this, we calculated the percentage of successful DNA amplifications for each factor (type of genetic marker, length of PCR fragments, and type of primer) in the references. In sequence, we compared the rate of fecal DNA amplification success between three different types of markers using fecal samples obtained from captive and wild goral populations. We also compared whether different types of primer sets and lengths of the amplified fragments could affect the DNA amplification success rate in captive gorals.
Previous experimental studies reporting differences in fecal DNA amplification success rate between different types of genetic markers (e.g., mitochondrial, microsatellite, and sex-linked), lengths of the amplified fragments (e.g., long, medium, and short), or types of primer sets (e.g., universal and specific) are rare (Byrne et al., 2021). Such a study would be a very useful starting point for conservation genetic experiments. The present review quantified and then compared different success rate of DNA amplification depending on various factors, including three markers, three lengths of amplified fragments, and two primer sets.
DNA Amplification Success of Three Different Markers
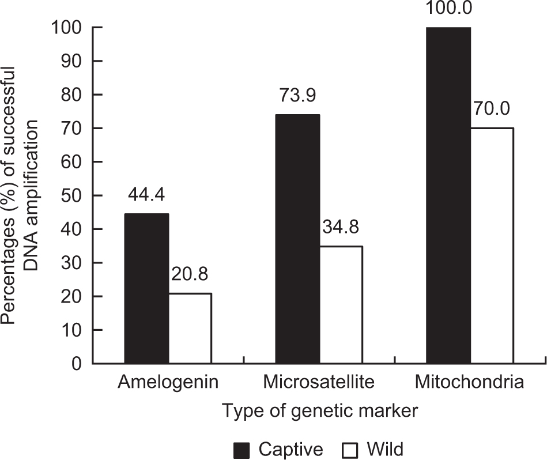
Three important papers reported that mitochondrial markers showed the highest percentage of DNA amplification success in both captive and wild goral populations, followed by microsatellite and sex-linked markers (Fig. 1). Large differences were seen between the two goral populations in the percentage of DNA amplification success for the three genetic markers. The captive population (24/24 = 100%) showed a higher mitochondrial marker PCR success rate (partial cytochrome b amplified by a primer set, NCF and NCR; Kim, 2021) than the wild population (28/40 = 70.0%). For microsatellite markers (n = 6 for each population; SY12A, SY50, SY58, SY129, SY17, and SY48), the captive population showed a 73.9% (17/23) PCR success rate but the wild population showed 34.8% (8/23; Kim, 2022). For the sex-linked marker (AMELX/Y amplified by a primer set, SE47 and SE48) between the two populations, the captive population (8/18 = 44.4%) showed a higher percentage of DNA amplification success than the wild population (5/24 = 21%) as microsatellite and mitochondrial markers (Kim et al., 2008).
Feces in wild populations are more easily influenced by various factors (e.g., heat, desiccation, and decomposition; Chame, 2003) than in captive populations, such as those in zoos or farms. Therefore, researchers can collect and use fresher feces from captive populations than from wild populations. More degraded samples are found in wild than in captive populations. These factors might affect the DNA amplification success rate of the fecal samples. Non-invasive samples contain mitochondrial DNA (e.g., cytochrome b gene and D-loop region) and nuclear DNA (e.g., microsatellites and sex-linked genes). Hundreds to thousands of copies of mitochondrial DNA are found in a cell, whereas only two copies of nuclear DNA are found in most cells (Birky et al., 1989). Thus, the rates of successful mitochondrial DNA extraction and amplification are higher than those of nuclear DNA (Frantzen et al., 1998; Kohn et al., 1999; Poole et al., 2001; Lucchini et al., 2002; Waits, 2004; Waits and Paetkau, 2005).
DNA Amplification Success According to Different Lengths of Amplified Fragments and Types of Primers
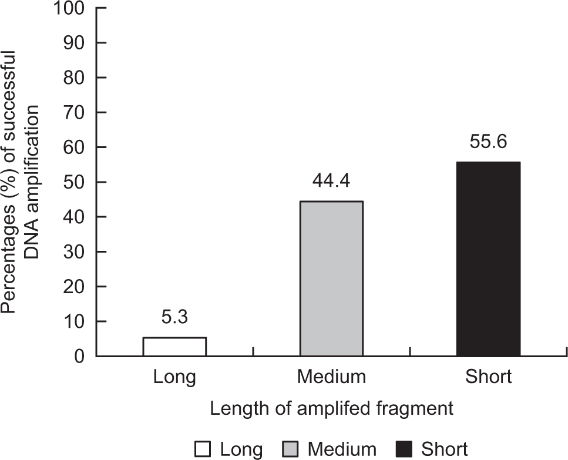
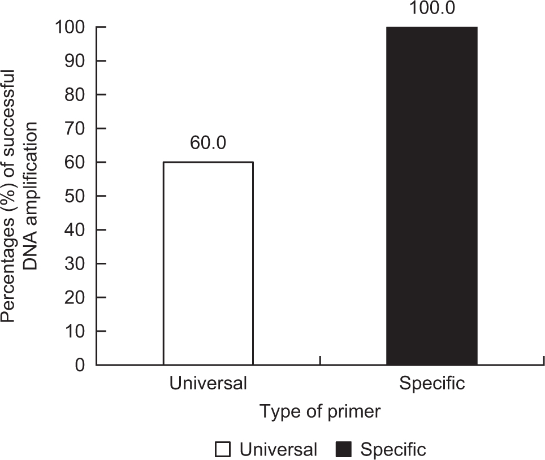
Regarding the percentage of DNA amplification success of sex-linked genes in the captive population according to different lengths of amplified fragments (e.g., long: ZFX/Y amplified by LGL331/335; and medium and short: AMELX/Y amplified by SE47/48 and SE47/53, respectively), the short-length fragment (10/18 = 55.6%) showed a higher value than the medium- (8/18 = 44.4%) and long-length fragments (1/19 = 5.3%; Fig. 2). In particular, the smallest value was below 10%, which is very low. Finally, two types of mitochondrial primers (e.g., specific: cytochrome b amplified by NCF/NCR; universal: cytochrome b amplified by L14841/H15149) showed different percentages of DNA amplification success in the captive population (Fig. 3). The goral-specific primers (100%) had much higher success rates than the universal primers (60.0%), although the length of the fragment amplified by NCF/NCR (378 bp) was slightly longer than that amplified by L14841/H15149 (376 bp, including primer regions).
Both the length of the amplified fragments and the type of primer set could be important factors affecting successful DNA amplification in non-invasive studies. Short fragments will be amplified better from low quantities of degraded DNA (Frantzen et al., 1998; Murphy et al., 2000; Roon et al., 2003). Sexing studies have been performed using various markers in mammals, such as SRY, ZFX/Y, and AMELX/Y. Among them, the AMELX/Y gene was frequently applied because of its short-length fragment (Yamamoto et al., 2002). The ZFX/Y primer set showed the lowest rate of successful sex-linked DNA amplification compared to other primer sets for AMELX/Y (SE47/48 and SE47/53). Waits and Paetkau (2005) previously reported that short target sequences are generally amplified more robustly. For species identification, two types of primers can be used in non-invasive studies, such as species-specific primers (Dalen et al., 2004) and universal primers (Kocher et al., 1989), designed using conserved regions in various species. The latter primers could be less efficient for DNA amplification success than the former primers due to a few nucleotides unmatched among different species.
Recommendation for Non-Invasive Conservation Genetic Experiments
In conclusion, the additional time, costs, and labor involved in working with fecal DNA should be considered in advance. This review showed that mitochondrial markers in both the captive and wild goral populations showed the highest percentage of success, followed by microsatellite and sex-linked markers. Fecal DNA amplification success rate was much higher in the captive than in the wild population due to relatively low DNA degradation. The DNA amplification success rate was much higher for shorter amplified fragments and a specific primer set than for longer amplified fragments and a non-specific primer set, respectively. Better results might be obtained by identifying and selecting the optimal amplified fragment length and type of primer set. This could be helpful as a first step for planning non-invasive genetic studies. We recommend beginning genetic experiments using mitochondrial markers and then using nuclear markers, such as microsatellite and sex-linked markers, to save time, costs, and labor. Also, we suggest that it is better to use fecal samples collected from a captive population in a pilot experiment and then use those collected from a wild population to decrease unnecessary experimental effort and increase experimental efficiency. Finally, it is expected that using short DNA markers rather than long DNA markers and specific markers rather than universal markers will yield better results in non-invasive conservation genetic studies.
Acknowledgments
This work was supported in part by the National Institute of Ecology, funded by the Ministry of Environment (MOE) of the Republic of Korea (No. NIE-B-2024-18). It was also partially supported by the Research Institute for Veterinary Science and the Brain Korea 21 Program for Veterinary Science, Seoul National University. We thank Professor H. Lee and Professor Y. J. Won for helping with conducting previous Korean goral studies and Dr. H. J. Bae for giving comments for this review paper.
Author Contributions
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Figures and Tables
Fig. 1
Percentages (%) of successful DNA amplification of the captive and wild populations for three different genetic markers.