Introduction
Unintentional dispersal of organisms has explosively increased; with recently increased human activities (Pimentel et al., 2005). Dispersed organisms that pose a major threat to the new environment or human health may be categorized as invasive species. These invasive species have been focused because they pose significant threats to biodiversity, environment, human health, and economy (Kim and Lee, 2019). However, it is difficult to predict and to estimate threats from newly introduced invasive species because detailed information about invasive species is not enough (Roques et al., 2009; Rowson et al., 2014; Bradshaw et al., 2016). Moreover, the newly introduced organism can adjust their behavior as well as physiological preference when it is introduced into new habitat (Berg et al., 2010). Therefore, eradication of invasive organism is almost impossible if the organism has successfully settled down in the new area. Only a small window is open to prevent successful settlement of invasive organism at the early stage of the introduction.
The leopard slug (Limax maximus) is known to have a distinctive body coloration similar to a leopard. It also has an extraordinarily large body length of ca. 15 cm. It has a pale brown or grey back with 4 ~6 interrupted dark bands and a mantle with spots (Barker, 1979). It originated in Europe. Its first description was conducted by Linnaeus in 1758. It is also well-known for its unusual mating behavior by a pair of slugs suspended by a mucus thread to exchange sperm (Karlin and Bacon, 1961). In its native habitat, this slug can serve as a detritivore that feeds on dead plant materials and fungi (Barker, 1999; Kozłowski, 2012).
Distribution of L. maximus in Europe, North Africa, and Asia has been reported. It has been introduced to other countries in North and South America, Africa, the Pacific Islands, Australia, Hawaii, and New Zealand (Barker and McGhie, 1984; Barker, 1999; Roth and Sadeghian, 2006; Joe and Daehler, 2008; McDonnell et al., 2009; Gaitán-Espitia et al. 2012). In Japan, this slug was confirmed in 2006 (Hasegawa et al., 2009). Based on this expansion of distribution, L. maximus is regarded as an invasive slug (Gaitán-Espitia et al., 2012). The preferred habitat of L. maximus includes forests, hedgerows, and gardens (Kerney and Cameron 1979; Cook and Radford, 1988; Meyer et al., 2013).
In Korea, L. maximus was detected at Bambat-Cheonggaeguri Park, Suwon in 2023 (Park et al., 2024). As it was recently introduced in Korea, it will not be difficult to eradicate this introduced slug with detailed ecological and physiological information of it. However, to make effective plans to eradicate this pest, its current dispersal status and possible dispersal area should be identified.
Before 2002, only two species (Limax, L. flavus Linnaeus 1758 and L. marginatus Müller, 1774) in genus Limax have been reported in Republic of Korea (Lee and Min, 2002). In the new habitat, this invasive herbivore species might affect plant species composition and diversity (Buschmann et al., 2005; Carlsson and Lacoursière 2005). In Hawaii and Japan, its food preference was tested.
To prepare for its potential ecological risk, possible food source of L. maximus was investigated by performing DNA meta-barcoding analyses of L. maximus feces. Feces were collected from L. maximus wild individuals and processed by meta-barcoding analyses to identify which plants were digested by these slugs. This information might give some clues to assume the potential environmental risk from L. maximus settlement on the Korean peninsula.
Materials and Methods
L. maximus habitat in Republic of Korea
Park et al., (2024) firstly reported L. maximus (Fig. 1) at Bambat-Cheonggaeguri Park (126.971138 E, 37.307072 N), Suwon, Gyeonggi-do in 2023 (Fig. 2). This park is surrounded by newly developed residential area in the south and forested area in the north including hobby farms, agricultural areas, and trails. Mt. Deokseong (160 m above sea level) is located ca. 1.5 Km away from the northern part of this park, and Mt.Gwanggyo (580 m above sea level) is located ca. 7 Km to the north-west side of this site. Therefore, this park has served as a buffer zone between natural environment and human residential area.
Sample collection
A total of 89 individuals of L. maximus (15 individuals during daytime and 74 individuals during night) (Fig. 2) were caught at the previously described site on September 19, 2023 (Table 1). These individuals were maintained in a plastic container (34 × 20.5 × 25 cm (H × W × L)) for one day without food source. From this plastic container, all feces were collected and transferred to 1.5 mL Eppendorf tubes. These collected feces were kept in four 1.5 mL Eppendorf tubes for ca. two months before DNA analysis. The collected sample was confirmed as L. maximus based on its morphological characters and barcoding analysis with 1,340 bp mitochondrial COI region (Nitz et al., 2009). Two stages of PCR were conducted for its molecular identification. The first fragment was amplified with mtCOI-1F-54 primer (5’-tttcaacaaaycataargatattgg-3’) and mtCOI-1R-53 primer (5’-aayaccaatagaaattatagcataaa-3’). The primer set for the second fragment included mtCOI-2F primer (5’-ttagcrggggcaattactatrc-3’) and mtCOI-2R primer (5’-cgaaaaca-gatattaacgaaccat-3’). Amplified sequences were checked by Sanger sequencing and combined to yield a 1,381 bp of mtCOI DNA sequence.
DNA meta-barcoding analyses
Fecal samples collected from L. maximus were thoroughly homogenization. After homogenized, 0.1 gram of each sample was transferred to a 2.0 mL tube with 3 mm zirconia ceramic beads for lysis with a Pre-Filled Bead Lysis Tube (Invirustech, Gwangju, Republic of Korea). DNA extraction was conducted using a Beniprep Soil/Fecal DNA Extraction Kit (IVT7003, Invirustech, Gwangju, Republic of Korea) according to the manufacturer’s protocol. The quantity and quality of extracted DNA were measured with an Epoch spectrophotometer (Agilent, Santa Clara, CA, USA).
For metagenomic analysis, Shotgun Metagenomics Sequencing was performed. Library preparation and quality control were conducted by Macrogen (Seoul, Republic of Korea). This sequencing generated 85,952,990 reads from fecal samples with a total read base of 13.0 Gbp. The GC content was 55.0%. This Shotgun Metagenomics Sequencing generated 93.1% of the Q30 data.
Pre-filtered reads were analyzed with Kraken2 suite (Lu et al., 2022). These reads were used as queries against the nt database using Kraken2. Analyzed results were summarized at the Streptophyta level to focus on possible food source of the leopard slug.
Results
The fecal sample generated 85,952,990 reads with a total read base of 13.0 Gbp. The GC content was 55.0%. The sequencing generated 93.1% of Q30 data. A total of 115K reads were identified as plants originated from this analysis. Among these, only Streptophyta, a terrestrial plant Phylum, was focused (Fig. 3). As a result, 22 families, 28 genera, and 26 species were identified as potential food sources of the leopard slug based on DNA meta-barcoding analysis.
At the species level, a total of 26 plant species were detected including not only economically important plants including cultivated squash (Cucurbita maxima), tomato (Solanum lycopersicum), black-eyed pea (Vigna unguiculata), and rice (Oryza sativa) but also environmentally important plants including pedunculate oak (Quercus robur), common ivy (Hedera helix), the tree of heaven (Ailanthus altissimus), and soft rush (Juncus effusus) (Table 2). Moreover, a wide range of plant species including cotton, chrysanthemum, clover, bitter cucumber, and blackcurrant were identified. For genus level identification, a total of 28 genera were identified (Table 3). Among the 28 identified genera, Solanum, Gossypium, Cucurbita, Ipomoea, and Brassica generally include diverse economically important crops.
Among 22 identified Families, Family Brassicaceae might be the major food source of L. maximus. Families Leguminosae, Solanaceae, Poaceae, Cucurbitaceae, and Fagaceae were also detected (Table 3).
Therefore, the settlement of this slug will affect not only agriculture but also natural environments. To reduce the ecological risk from this slug, more detailed information about microhabitats as well as the food preferences of L. maximus in Republic of Korea might be required.
Discussion
Potential food sources of the leopard slug, an invasive species, in Republic of Korea were investigated by DNA meta-barcoding analysis. Feces of wild individuals of L. maximus were collected for this analysis. From this analysis, various plant species were identified as potential food sources of L. maximus in Republic of Korea. Komatsu and Saeki (2022) have also reported that 26 plant species are possible food sources of L. maximus based on barcoding analysis in Japan. However, there was no common plant species detected between Japan and Republic of Korea. This difference might be caused by different flora in these two countries that are close in distance but separated by ocean. These results suggest that L. maximus can feed on a broad range of plant species.
In this study, L. maximus was found at a park that is highly connected to human activities such as walking with pets and at an agricultural area that is used for pumpkins, tomatoes, beans, rice, and other crops (Table 1). Many slugs can be a disease vector as an intermediate or final host for diverse parasitic nematodes and microorganisms causing various diseases in humans as well as other mammals (Wang et al., 2008; Sudhaus, 2018). It was reported that there was a parasite infection risk when L. maximus was handled by naked hands in China and an angiostrongyliasis was reported in a young man who ingested L. maximus in a garden in Australia (Senanayake et al., 2003; Li et al., 2024). Angiostronyliasis is known as a Class 4 infectious disease in Republic of Korea, which might be caused by eating raw or undercooked snails or slugs contaminated with parasites. Angiostrongyliasis could eventually lead to death or permanent brain and nerve damage in human (Wang et al., 2008). In cases affecting dogs, Fuehrer et al. (2020) reported that Crenosoma vulpis was detected in L. maximus and C. vulpis is a well-known parasite that could affect the lung and respiratory tract of dogs. It could cause serious respiratory diseases that could be fatal. Among diverse canines in Korea, foxes are the most popular host for C. vulpis, but C. vulpis infection had been reported in pet dogs (Choi et al., 2014). Therefore, more attention is required when waling with pets in Suwon, where L. maximus, a potential host for C. vulpis, was detected.
As a newly introduced invasive species, L. maximus can have diverse impacts on agricultural and natural environment as well as human and pet health. Therefore, further study is needed to perform risk assessment for this slug because of the broad spectrum of its impacts. In addition, a focused control measure will be able to suppress the high density of this slug population before it is extensively dispersed as an early stage of the introduction of this slug.
Author Contributions
The authors confirm contribution to the paper as follows: study conception and design: HK, YP, YC; data collection: YP; analysis and interpretation of results: HK and KP; draft manuscript preparation: HK, KP, YP, YC. All authors reviewed the results and approved the final version of the manuscript.
Figures and Tables

Limax maximus at its natural habitat in Republic of Korea. (A) Overview of the L. maximus habitat in Suwon, (B) Walkin with a dog at the L. maximus habitat in Suwon, (C) L. maximus found in its natural habitat.
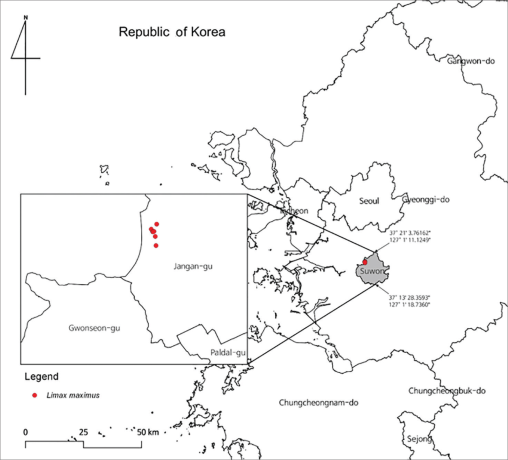
The field site of collecting L. maximus in Republic of Korea. Red dots indicate where L. maximus were collected.
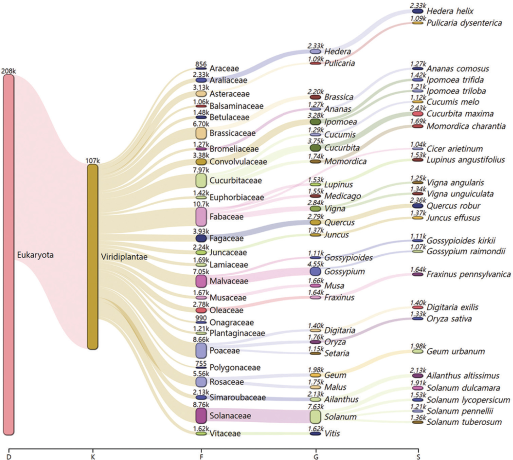
Sankey visualization of barcoding results focused on Phylum Streptophyta. At the species level, 26 Species, 28 Genera, and 22 Families were identified with more than 1,000 reads after aligned as descending order of all identified sequences. (D = Domain, K = Kingdom, F = Family, G = Genus, S = Species).
