Introduction
According to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC), a 4°C increase in global atmospheric temperature is projected by 2100 (Kikstra et al., 2022). The effects of global climate change, including sea-level rise, droughts, and floods, result in severe and widespread damage to the ecosystem. A global average temperature rise of 1.5-2.5°C could lead to the extinction of approximately 20-30% of animal and plant species, with more than 40% of species potentially facing extinction if temperatures rise by 4°C (IPCC, 2014). Thus, research aimed at protecting biodiversity is critical, especially in areas susceptible to climate change such as islands, coasts, and subalpine regions (Hardy, 2003). Plant species in subalpine regions are particularly vulnerable to extinction due to limited opportunities for upslope migration (Bell et al., 2014; Koo et al., 2017; Randin et al., 2009).
Korean fir (Abies koreana), endemic to Korea, is found on the summits of Mts. Halla, Jiri, Mudung, Kaji, and Deogyu in the southern part of the Korean peninsula (Lee, 1982). Due to its declining population from climate change, A. koreana is classified as a threatened species by the International Union for Conservation of Natural Resources (IUCN) and is designated as a biological indicator of climate change in Korea (Kim et al., 2011; Lee et al., 2010). The regional decline of the A. koreana population has been occurring since the 1980s, driven by poor growth and dieback due to global warming, along with various biotic and abiotic stresses such as warm winters and droughts (Koo et al., 2001; Woo, 2009; Lim et al., 2006; Woo et al., 2008). Particularly, it has been reported that A. koreana in the Youngsil region is declining due to an insufficient water supply compared with the Nambyuk region of Mt. Halla (Kim et al., 2017). Therefore, extensive research is necessary for the conservation and effective forest management of A. koreana.
We recently conducted de novo RNA sequencing (RNA-seq) on A. koreana trees that were exposed to elevated CO2 levels and heat, in an effort to elucidate the transcriptomic alterations associated with environmental stress (Hwang et al., 2018; 2019). These findings have significantly enhanced our comprehension of the molecular responses and environmental stress response pathways in A. koreana when subjected to elevated CO2 and heat. The various genes identified from the de novo RNA-seq data were evaluated as candidate marker genes in the ecologically stable and vulnerable regions of Mts. Halla and Jiri (Kim et al., 2020). Moreover, molecular marker genes have been used as environmental risk assessment tools in LM cotton to protect ecosystem safety and monitor of bifenazate resistance in two-spotted spider mites, Tetranychus urticae (Jo et al., 2016; Lee et al., 2011).
We previously performed a comprehensive transcriptomic analysis of A. koreana under drought stress and identified four highly expressed genes (Akoreana1SL019192t0002, Akoreana1SL009606t0001, Akoreana1SL022564t0001, and Akoreana1SL003793t0001). In this study, we assessed the expression levels of these four genes using quantitative real-time polymerase chain reaction (qRT-PCR) in 10 samples of A. koreana collected from both the ecologically stable (Nambyuk) and vulnerable (Youngsil) regions of Mt. Halla. The results of this study will facilitate the development of diagnostic markers for assessing the vulnerability of A. koreana to environmental stress, as well as the development of strategies for conservation and restoration of the subalpine ecosystem amid climate change.
Materials and Methods
Sample collection
The needles (leaves) of A. koreana plants were collected from both the ecologically stable (Nambyuk) and vulnerable (Youngsil) regions of Mt. Halla (Fig. 1), each located approximately 1,500 m from the summit. Plants were collected from 10 sites per region and immediately frozen in liquid nitrogen. The samples were then transported to the laboratory and stored at –80°C. Subsequently, total RNA was extracted using the Ribospin Seed/Fruit RNA mini kit (GeneAll, Seoul, Korea) according to the manufacturer’s instructions.
Analysis of drought stress-responsive genes in A. koreana
In a previous study, drought stress-responsive genes in A. koreana were isolated using comparative RNA-seq analysis (Park et al., 2018). Briefly, raw reads were trimmed with Trimmomatic (version 0.32), and then assembled de novo using Trinity software (version R20140717), with default settings, to generate a suitable set of reference contigs named unigenes (Grabherr et al., 2011). These unigenes were functionally annotated with BLASTX (BLAST 2.6.0+), using the NCBI non-redundant protein sequence database. Furthermore, gene expression profiles from the RNA-seq data were analyzed with RNA-Seq by Expectation-Maximization (RSEM) software, part of the Trinity package (Schmieder & Edwards, 2011). Lastly, drought stress-responsive genes showing more than a 2-fold increase in expression (p-value ≤ 0.05) were selected for further analysis.
Gene expression analysis by qRT-PCR
A. koreana cDNA was synthesized from 1 μg of total RNA using the ReverTra Ace-a kit (Toyobo, Osaka, Japan), following the manufacturer’s instructions. Expression levels of drought stress-responsive genes and AkActin (reference gene) were quantified by qRT-PCR on the CFX96 real-time system using IQTM SYBR Green Supermix, processed with the Bio-Rad CFX Manage program (Bio-Rad, Hercules, USA) (Lee et al., 2023). Each qRT-PCR reaction comprised 1 μl of diluted cDNA, 10 μl of 2× IQ SYBR Green Supermix, 1 μl each of 10 pM forward and reverse primers, and 7 μl of H2O. The qRT-PCR conditions included an initial step at 95°C for 10 min, followed by 55 cycles at 95°C for 15 s, 55°C for 15 s, and 75°C for 30 s. Primer sequences used for qRT-PCR are detailed in Table 3. The expression levels of drought stress-responsive genes were normalized against AkActin using the comparative CT (ΔΔCT) method (Livak & Schmittgen, 2001). Error bars indicate standard deviation values for triplicates.
Results and Discussion
Selection of A. koreana sampling sites on Mt. Halla
A. koreana exhibits the widest distribution on Mt. Halla, and its decline has been the most severe across the Korean peninsula. The decline in A. koreana population is likely due to complex interactions among various environmental factors induced by global warming (Drohan et al., 2002; Duchesne et al., 2005). The population of A. koreana in the Youngsil region of Mt. Halla decreased by 21.5% (25.3 ha) compared to 2006 (Kim et al., 2017). For this study, two regions on Mt. Halla were selected for sample collection: Nambyuk (NB) and Youngsil (YS) (Fig. 1). NB, situated south of the peak of Mt. Halla, supports ecologically stable growth of A. koreana. In contrast, YS is located southwest of the Mt. Halla peak and designated as an ecologically vulnerable region because of the declining A. koreana population. Ten A. koreana plants were sampled from each region (Fig. 2), with the geographic coordinates as follows: NB: 33.358731-33.358936 (latitude) and 126.508706-126.508871 (longitude); YS: 33.357776-33.357923 (latitude) and 126.523485-126.523713 (longitude) (Table 1). These plants were then subjected to gene expression analysis to identify the ecologically stable and vulnerable regions of A. koreana in Mt. Halla.
Isolation and characterization of drought stress-responsive genes in A. koreana
A. koreana, an endangered species sensitive to drought, is experiencing severe decline due to global warming. Therefore, it is crucial to uncover the adaptive molecular mechanisms utilized by A. koreana in response to drought stress. We previously conducted a comprehensive transcriptomic analysis on A. koreana exposed to varying periods of drought stress and identified genes that were upregulated and downregulated in comparison with untreated (control) plants (Park et al., 2018). In this study, we focused on genes whose expression increased by more than 2-fold under drought stress. Among these, four genes (Akoreana1SL019192t0002, Akoreana1SL009606t0001, Akoreana1SL022564t0001, and Akoreana1SL003793t0001) were selected for further analysis. Gene annotation with BLASTX (BLAST 2.6.0+) showed that Akoreana1SL019192t0002 codes for Myb domain protein 21, which contributes to salt tolerance in Arabidopsis thaliana (Zhang et al., 2021) and Myb-related protein 305 in Populus trichocarpa; Akoreana1SL009606t0001 encodes ATP-binding cassette 14, which strengthens the abscisic acid signaling in guard cells and enhances water use efficiency in Arabidopsis (Kuromori et al., 2016) and ABC transporter G family member 21 isoform X1 in Populus; Akoreana1SL022564t0001 encodes ethylene-responsive element binding factor 3 (ERF3), associated with drought stress responses in Arabidopsis (Song et al., 2005) and ERF9 in Populus; and Akoreana1SL003793t0001 encodes a basic helix-loop-helix (bHLH) DNA-binding superfamily protein, activated by the presence of NaCl or glucose in Arabidopsis (Ikeya et al., 2020) and a bHLH family protein in Picea sitchensis (Table 2). Subsequently, the expression levels of these four genes were validated in the sampled A koreana plants through qRT-PCR using gene-specific primers (Table 3).
Increased expression of drought stress-responsive genes in the ecologically sensitive area of Mt. Halla
Expression levels of the four genes were examined by qRT-PCR in 10 A. koreana samples collected from each of the ecologically stable (Nambyuk, NB-1-10) and vulnerable (Youngsil, YS-1-10) regions of Mt. Halla (Fig. 3). All four genes exhibited higher expression levels in YS samples compared to NB samples (Fig. 3). Specifically, Akoreana1SL019192t0002 demonstrated approximately 100-3,300-fold higher expression in YS samples than in NB samples. Similarly, Akoreana1SL009606t0001, Akoreana1SL022564t0001, and Akoreana1SL003793t0001 showed roughly 11-106-, 4.3-17-, and 8-27-fold higher expression in YS samples compared to NB samples, respectively. Collectively, these results suggest that the four genes could serve as diagnostic markers for assessing the ecologically stable and vulnerable regions of Mt. Halla.
In conclusion, global warming poses a significant threat to the biodiversity of natural ecosystems, particularly in vulnerable regions like subalpine forests. The population of A. koreana, endemic to subalpine regions in southern Korea, continues to decline due to warm winter temperatures and water stress caused by global warming. Therefore, it’s crucial to investigate the mechanisms of environmental stress resistance in A. koreana under field conditions. Our findings indicate that the diagnostic genes can effectively assess the ecological vulnerability of A. koreana under various environmental stresses and aid in conservation efforts in subalpine ecosystems. Additionally, this study lays a strong foundation for further molecular studies and the development of conservation and restoration strategies for these ecosystems.
Acknowledgments
We thank Dae Young Jeong and Jong-won Park for their assistance in sample collection from Mt. Halla and in conducting the qRT-PCR experiments. This work was supported by a grant from the National Institute of Ecology (NIE), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIE-B-2024-15).
Author Contributions
H.C.P. designed the experiments. H.C.P., D.Y.P and D.Y.L. conducted the experiments and analyzed the data. H.C.P. drafted the manuscript and maintained the plant materials. H.C.P and D.Y.P discussed the results and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Figures and Tables
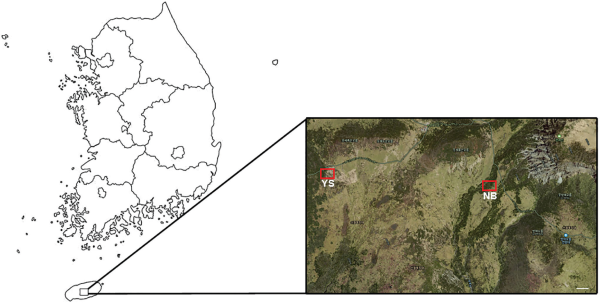
Area of investigation of Abies koreana in the subalpine region of Mt. Halla in Korea. Red rectangles denote the ecologically stable region (Nambyuk, NB) and vulnerable region (Youngsil, YS), classified based on the growth of A. koreana. Scale bar = 100 m.

Sampling sites of A. koreana in the ecologically stable region (Nambyuk, NB) and the vulnerable region (Youngsil, YS) of Mt. Halla. (A) Depicts the locations of 10 sampling sites in NB. (B) Shows a photograph of NB. (C) Illustrates the locations of 10 sampling sites in YS. (D) Displays a photograph of YS. Scale bars represent 10 m.
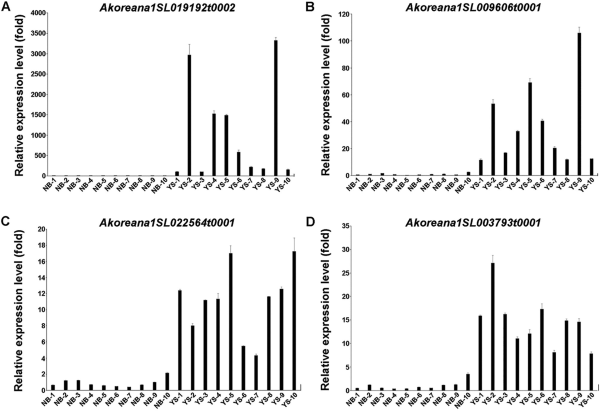
Expression analysis of four genes in the ecologically stable (Nambyuk, NB-1-10) and vulnerable (Youngsil, YS-1-10) regions of A. koreana in Mt. Halla Samples were taken from 10 sites each in the ecologically stable (Nambyuk, NB) and vulnerable (Youngsil, YS) regions of Mt. Halla. (A-D) Show the expression levels of Akoreana1SL019192t0002 (A), Akoreana1SL009606t0001 (B), Akoreana1SL022564t0001 (C), and Akoreana1SL003793t0001 (D).

Geographic coordinates of A. koreana sampling sites in the ecologically stable region (Nambyuk, NB) and the vulnerable region (Youngsil, YS) of Mt. Halla