ISSN : 2765-2203
ISSN : 2765-2203
Vol.5 No.4

Abstract
Butterflies are insects that consistently attract significant attention due to their beautiful appearance. In this review, we analyzed the feeding preferences of endangered butterfly larval host plants and adult nectar plants. We examined host and nectar plants of all Korean endangered butterfly species by referring to previous literature. Each endangered butterfly species in this review exhibited a narrow range of feeding preferences, utilizing between 0 to 3 plant families and 0 to 14 plant species as host plants. Both Aporia crataegi and Melitaea ambigua had the highest number of host plant families (n = 3), and A. crataegi had the highest number of host plant species (n = 14). In total, 13 families and 42 species of host plants were identified as being utilized by 14 target endangered butterfly species. Conversely, each endangered butterfly species in this review demonstrated a broad range of feeding preferences, utilizing between 2 to 12 plant families and 2 to 21 plant species as nectar plants. M. ambigua had the highest number of nectar plant families (n = 12) and the highest number of nectar plant species (n = 21). In total, 21 families and 61 species of nectar plants were identified as being utilized by 14 target endangered butterfly species. This review is the first to comprehensively summarize the host and nectar plants of all endangered butterflies in South Korea and could serve to establish future restoration plans for these butterflies.


Abstract
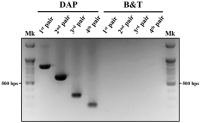
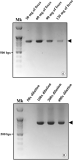
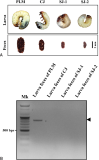
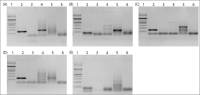
Polyphylla laticollis manchurica (PLM), distributed across four regions along the Geumgang River in South Korea, is classified as endangered. Its developmental cycle from larva to adult spans 3-4 years, making it challenging to verify its presence or absence within its habitat, except during the adult phase. Over the last two decades, multiple noninvasive genomic DNA (gDNA) sampling methods, including ones using eggshells, feces, mucus, and molted skin, have been developed. These methods facilitate the identification of species and genetic analysis without causing harm to the target species. This study considers the highly conserved cytochrome b sequence region of PLM for PCR amplification using a direct gDNA extraction method with feces. To establish an effective direct gDNA extraction method, this study used DAP buffer – previously proven effective for extracting gDNA from feces and mucus of invertebrates – and compared PCR amplification efficiency based on feces weight and dilution factor. Optimal PCR amplification efficiency was achieved using 10 mg of feces per 100 μL DAP buffer and a 200-fold dilution. Species-specific PLM markers distinguish PLM from sympatric species within their habitats. The noninvasive species identification method employing feces permits rapid species identification by circumventing the sequencing process. This rapid identification technique could serve as a preliminary monitoring tool for the protection of the endangered PLM’s habitat when adult individuals are not visible.

Abstract
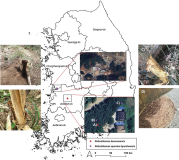
Drought stress poses a significant threat to ecosystems, particularly affecting the subalpine ecosystem most severely. Korean fir (Abies koreana) is found in the subalpine region of Korea, including the summits of Mts. Halla, Jiri, Mudung, Kaji, and Deogyu. Although A. koreana is recognized as a species sensitive to drought stress by the Korean government, its response mechanisms are poorly understood. We previously conducted a comprehensive transcriptomic analysis of A. koreana under drought conditions. In our current study, we selected four genes exhibiting high expression, namely Akoreana1SL019192t0002, Akoreana1SL009606t0001, Akoreana1SL022564t0001, and Akoreana1SL003793t0001. Additionally, A. koreana samples from ten locations each in the ecologically stable (Nambyuk) and vulnerable (Youngsil) areas of Mt. Halla were analyzed using quantitative real-time polymerase chain reaction to measure gene expression. All four genes showed higher expression levels in samples from the vulnerable area compared to those from the stable area. These findings suggest that these genes could serve as diagnostic markers to assess A. koreana’s vulnerability to drought stress in natural settings.



Abstract
Invasive alien species, along with climate change, are major contributing factors to biodiversity loss, and their unintentional introduction through imported goods is increasing. Termites, as wood-feeding insects, pose a significant threat when introduced into Korea via imported wood, necessitating a rapid and accurate identification method. In this study, we developed a diagnostic method based on species-specific genetic sequences for identifying Reticulitermes kanmonensis, a recently identified species. Termite samples were collected from Wanju-gun, Jeollabuk-do, and subjected to whole-genome sequencing to pinpoint species-specific genetic sequences. Utilizing these sequences, we designed primer sets and employed TaqMan-based primer sets and qPCR analysis to select the final primer sets capable of rapidly distinguishing R. kanmonensis. The genetic detection method developed here offers a rapid means of identifying alien termite species, likely enhancing termite management and quarantine practices in Korea.



Abstract
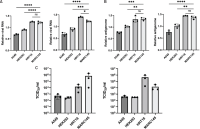
Viruses mutate and form quasispecies within the host, from which the next generation is selected. Studying the selected mutations of an animal virus across different host species can aid in predicting how the virus might evolve to manage future potential pandemics in humans. In this study, both high-passaged (p44) and low-passaged (p4) Shaan viruses of bat origin in MARC-145 cell lines, derived from African green monkey kidney, were passaged once into human A549, HEK-293, and HRT-18 cell lines as different host species models. High-throughput sequencing data showed that there were distinct selected mutations and single nucleotide variants (SNVs) between the low- and high-passaged Shaan viruses in MARC-145 cells. After host switching, these mutation patterns were consistently observed among the same host cells in triplicate experiments, suggesting a host cell-specific selection pattern for the progeny of Shaan viruses. Notably, HRT-18 cells of colorectal adenocarcinoma origin produced more unique selected mutations and unique SNVs. While Shaan virus-specific transcripts associated with the N gene were most abundant in MARC-145, A549, and HEK-293 cells, those associated with the M gene were most abundant in HRT-18 cells. After host switching, the relative viral RNA levels in the HRT-18 cells were significantly higher than in A549 and HEK-293 cells. Thus, this study provides evidence of host-specific mutation patterns by cell type and host cells in the evolution of Shaan virus.






Abstract
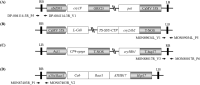
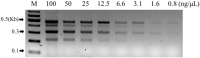
This study focuses on developing a multiplex PCR assay for the simultaneous detection of four specific living modified (LM) maize varieties (DP-004114-3, MON89034, MON88017, and MON87403) approved for use in South Korea. The increasing use of LMO maize in food, feed, and industrial applications has heightened concerns about unintentional environmental releases, underscoring the need for reliable detection methods. Genetic information for each LMO event was obtained, and event-specific primers were designed to ensure accurate amplification. The developed multiplex PCR method underwent rigorous validation through specificity assays, analysis of certified reference material (CRM) mixtures, and limit of detection (LOD) testing. The assay effectively detected all target events with high sensitivity, capable of identifying as little as 12.5 ng μL–1 of genomic DNA. Furthermore, this method was utilized with suspicious samples gathered during the monitoring of unintentional LMO releases in natural settings, confirming its utility in practical applications. Our results show that the multiplex PCR assay is a robust and efficient method for the detection and monitoring of LM maize in both environmental and regulatory contexts. This technique holds considerable potential to safeguard natural ecosystems by preventing unintentional LMO releases and ensuring adherence to biosafety regulations.




Abstract
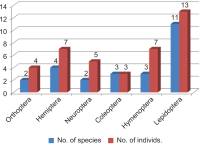
We conducted a monitoring study on hitchhiker insect pests aboard international vessels entering Korea in 2023, where we collected 562 individuals from 247 vessels. Of these, 512 individuals were identified as 260 species across 66 families in eight orders, while the remaining 50 individuals were classified at the family level. From the survey of the distribution of these 260 species (512 individuals) identified at the species level, 25 species (39 individuals) across 18 families of six orders were recognized as not-distributed species in Korea. This group included three regulated species (five individuals), Oryctes rhinoceros (Scarabaeidae, Coleoptera), Oecophylla smaragdina (Formicidae, Hymenoptera), and Solenopsis invicta (Formicidae, Hymenoptera) which are listed by the Animal and Plant Quarantine Agency of Korea. Accordingly, continuous quarantine inspections on vessels from native regions are necessary to prevent the entry of these 25 non-distributed species confirmed in this study, in addition to enhancing monitoring around domestic port entry points. As part of the quarantine inspection, we have provided detection information on these 25 non-distributed species in Korea.