Introduction
Invasive alien species, in conjunction with climate change, are among the five crucial contributors to biodiversity losses, with unintentional introductions through imported goods rising globally (IPBES, 2023). In 2023, South Korea reported exports of USD 632.2 billion and imports of USD 642.6 billion, marking an increase of 10-20% over the past decade (Biswas, 2023; MOTIE, 2024), thereby heightening the risk of unintentional introduction of alien species. The import of large quantities of agricultural products and raw materials such as timber, which likely introduce wood-feeding insects like termites, has particularly increased (Kim & Lee, 2019; Eyer & Vargo, 2021). Termites play a crucial role as decomposers in ecosystems by breaking down plant materials, yet they also cause significant economic damage to wooden structures, leading to annual losses of $40 billion (Rust & Su, 2012).
In South Korea, concerning the introduction of alien termite species, six ant species from five families and five genera were reported to have been quarantined 67 times in various wood materials and other plants imported from 16 countries between 2003 and 2022; Coptotermes acinaciformis and Porotermes quadricollis were first identified in 2007, followed by Coptotermes formosanus in 2010, Coptotermes curvignathus in 2017, and Coptotermes gestroi in 2020. These species are targeted as controlled pests in South Korea. However, there are 19 quarantine stations at 22 ports across the country, and cases of quarantine involving alien termite species continue to occur, indicating potential nationwide introductions (Kim & Kim, 2024).
Rapid identification of alien termite species (introductions) is crucial for effective pest control; however, taxonomic studies have been insufficiently conducted with only 15 termite cases identified at the species level out of a total of 67 (Kim & Kim, 2024). Cryptotermes domesticus and Incisitermes minor were discovered in May and September in Gangnam-gu, Seoul, and Changwon-si, Gyeongsangnam-do, South Korea, respectively. It took one week to genetically identify the alien species introduction status of R. speratus, which was found in Gangnam-gu. (ME, 2023a, 2023b, 2023c).
This study aims to propose a diagnostic method using species-specific genetic sequences for Reticulitermes kanmonensis, a native species in South Korea, to accurately and swiftly determine whether termites found in imported wood are invasive species.
Materials and Method
Sample collection of Reticulitermes speratus and Reticulitermes kanmonensis
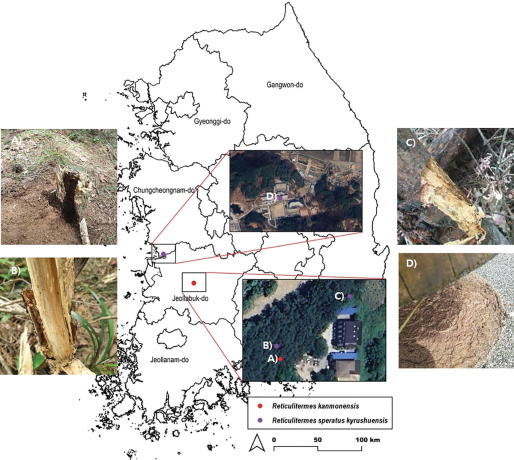
To design a diagnostic method based on species-specific genetic sequences for R. kanmonensis, the native species R. speratus was collected and used as a control for comparative analysis. Reticulitermes speratus was collected in June and August 2023 from two locations: Wanju-gun in Jeollabuk-do, where R. kanmonensis was first identified in Seocheon-gun in Chungcheongnam-do, South Korea (Lee et al., 2015). R. speratus samples were sourced from both standing and fallen dead pine trees or stumps using an axe; R. speratus specimens collected from a single stump and tree were treated as a single sample and preserved in 95% ethanol. Four samples, each comprising a minimum of one army ant and two worker ants, were collected and utilized (Fig. 1).
Sequencing
The Clear-S Quick DNA extraction kit (IVT-3002, Invirustech, S. Korea) was used to extract DNA from the collected termite samples according to the manufacturer’s protocol.
Production of primers for gene amplification and selection
To design primers for species amplification, this study identified candidate sequences from WGS analysis of genomic DNA. Subsequently, Primer3 software (Untergasser et al., 2012) was employed to design five pairs of primer and probe sets for each species (Table 1), focusing on the precise amplification of species-specific genetic sequences. The properties of these primers were optimized considering Tm values, GC content, and potential secondary structure formation.
qPCR was conducted using extracted and refined genes from two species as templates. The conditions were as follows: 10 μL EzAmpTM HS qPCR 2X Master Mix (SYBR Green, Low Rox), 3 μL (1 ng/μL) of genomic DNA, 0.5 μL of each primer at 10 pmol, and 6 μL of nuclease-free water for a final volume of 20 μL. The cycling condition commenced with an initial denaturation at 95°C for five minutes, followed by 45 cycles of denaturation at 95°C for 10 seconds and annealing/extension at 60°C for 30 seconds. Melting curve analysis was performed, increasing the temperature by 0.5°C/s from 60°C to 95°C. Two primer sets were selected for each species based on their PCR amplification performance and specificity.
Results and Discussion
Whole-genome sequencing (WGS)
WGS was conducted using the Illumina Novaseq platform with 150 bp libraries in a paired-end sequencing format, generating each 150 bp read. This sequencing produced 282,273,060 RAW reads with Q20 (97.09%), Q30 (92.39%), and GC content (46%). WGS data for other termite species were obtained from the SRA database of NCBI GenBank. WGS data estimated the total genome size of R. kanmonensis to be approximately 270-277 Mb, of which about 81% was repetitive sequences.
For the effectiveness of the designed primers, it is crucial that they exhibit a low mutation rate. HMMER and BLASTn analyses of the designed primers and amplified sequences revealed that all five sets showed high similarity to protein-coding regions of other termites, suggesting these as potential protein-coding regions of R. kanmonensis (Tables 2 and 3). Nevertheless, some primer sets did not align with specific protein-coding regions, indicating the need for further research, including a full genome analysis of R. kanmonensis.
Development and selection of species-specific primers
A TaqMan-based probe method, known for its high specificity and sensitivity, was employed to enhance accuracy. DNA was extracted from each termite, followed by WGS to identify species-specific sequences. Subsequently, PCR results were confirmed, and target sequences were found to be well-amplified (Table 4). By selecting and verifying primer sets that efficiently amplify only the target species, R. Kan set 1 and R. Kan set 4 were chosen as species-specific primers for R. kanmonensis due to their high amplification efficiency and lack of non-specific bands (Fig. 2).
Specificity and sensitivity of species-specific primers
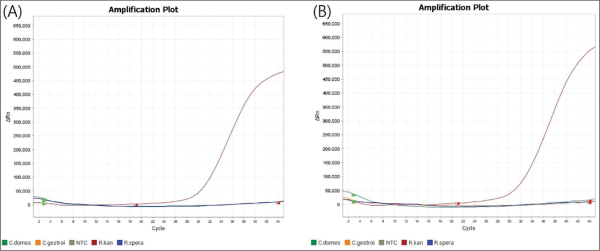
In qPCR, the Ct value, representing the Cycle Threshold, is used to estimate the initial amount of target DNA. Given the higher amplification sensitivity, even a low initial concentration of target DNA can be detected, resulting in a lower Ct value. To confirm the sensitivity of species-specific primers, the Ct value of each primer set was determined. The primer set with the lowest Ct value was selected as the species-specific marker.
Although the initially selected primer set was adequate to obtain results, the Ct values were compared to ensure more reliable and accurate outcomes (Fig. 3). In the case of R. kanmonensis, the Ct value for R. Kan set 1 was 30.945, indicating higher sensitivity compared to R. Kan set 4 (Table 5). Further analysis of PCR gel loading results (Fig. 2) showed that in R. kanmonensis, R. Kan set 1 amplified more specifically than R. Kan set 4, suggesting that R. Kan set 1 is likely more effective.
Author Contributions
Conceptualization by Soon Jae Eum and Kibeom Park, Research and data collection by Youngjun Park and Youngho Cho.
References
ME (2023a, last modified 2023.5.19) Ministry of Environment Post about termites, Ministry of Environment of Korea, https://www.me.go.kr/home/web/board/read.do?pagerOffset=0&maxPageItems=10&maxIndexPages=10&searchKey=title&searchValue=%ED%9D%B0%EA%B0%9C%EB%AF%B8&menuId=10525&orgCd=&boardId=1601930&boardMasterId=1&boardCategoryId=&decorator, accessed, 2024.08.30
ME (2023b, last modified 2023.5.24) Ministry of Environment Post about termite, Ministry of Environment, https://www.me.go.kr/home/web/board/read.do?pagerOffset=0&maxPageItems=10&maxIndexPages=10&searchKey=title&searchValue=%ED%9D%B0%EA%B0%9C%EB%AF%B8&menuId=10525&orgCd=&boardId=1602970&boardMasterId=1&boardCategoryId=&decorator, accessed, 2024.08.30
ME (2023c, last modified 2023.9.26) Ministry of Environment Post about termite, Ministry of Environment, https://www.me.go.kr/home/web/board/read.do?menuId=10525&boardMasterId=1&boardCategoryId=39&boardId=1626430, accessed, 2024.08.30
MOTIE (2024, last modified 2023.12.31) Ministry of Trade, Industry and Energy https://www.data.go.kr/data/3039964/fileData.do, accessed, 2024.08.30
Figures and Tables
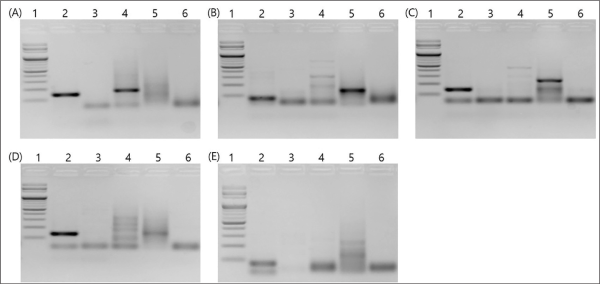
Amplification Results Using Primer Sets for the Specific Detection of R. kanmonensis. Lane 1: 100 bp Marker; lane 2: R. kanmonensis DNA; lane 3: R. speratus DNA; lane 4: C. domesticus DNA; lane 5: C. gestroi DNA; lane 6: Distilled Water (D.W). Set designations: (A) R. Kan Set 1, (B) R. Kan Set 2, (C) R. Kan Set 3, (D) R. Kan Set 4, (E) R. Kan Set 5.

Coding protein information of genetic locations for species-specific primer sets of Reticulitermes kanmonensis

Sequence of PCR amplicons from species-specific primer sets for native termite species in Korea: Reticulitermes kanmonensis