4권 2호

초록
The Kentish Plover (Charadrius alexandrinus; family Charadriidae; genus Charadrius) is a small bird that moves from continent to continent depending on the season. On the Kentish Plovers, phylogenetic studies have been widely conducted to classify different species or subspecies and to determine the time of speciation. However, the perspectives on the interspecific or intraspecific relationships in the phylogenetic analysis of Kentish Plovers remain debatable. Here, we reviewed the differences between the Kentish and Snowy Plovers (C. nivosus) in terms of their morphology, ecology, and genetic information. Particularly, their differences in genetic information can be well demonstrated; however, the intraspecies differences in the populations that live in different environments can relatively be poorly explained. We suggest that not only genetic features but also morphological, ecological, and behavioral traits are important when comparing the Kentish Plovers with other species, such as the Snowy Plovers, in phylogenetic studies. Furthermore, we suggest that phylogenetic studies on the subspecies of the Kentish and Snowy Plovers should be conducted for their better identification.
Abstract
The Kentish Plover (Charadrius alexandrinus; family Charadriidae; genus Charadrius) is a small bird that moves from continent to continent depending on the season. On the Kentish Plovers, phylogenetic studies have been widely conducted to classify different species or subspecies and to determine the time of speciation. However, the perspectives on the interspecific or intraspecific relationships in the phylogenetic analysis of Kentish Plovers remain debatable. Here, we reviewed the differences between the Kentish and Snowy Plovers (C. nivosus) in terms of their morphology, ecology, and genetic information. Particularly, their differences in genetic information can be well demonstrated; however, the intraspecies differences in the populations that live in different environments can relatively be poorly explained. We suggest that not only genetic features but also morphological, ecological, and behavioral traits are important when comparing the Kentish Plovers with other species, such as the Snowy Plovers, in phylogenetic studies. Furthermore, we suggest that phylogenetic studies on the subspecies of the Kentish and Snowy Plovers should be conducted for their better identification.

초록
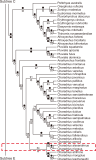
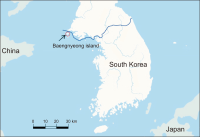
The Korean field mouse, Apodemus peninsulae mitochondrial genome has previously been reported for mice obtained from mainland Korea and China. In this investigation the complete mitochondrial genome sequence for a mouse obtained from Baengnyeong Island (BI) in South Korea was determined using high-throughput whole-genome sequencing for the first time. The circular genome was determined to be 16,268 bp in length. It was found to be composed of a typical complement gene that encodes 13 protein subunits of enzymes involved in oxidative phosphorylation, two ribosomal RNAs, 22 transfer RNAs, and one control region. Phylogenetic analysis involved 13 amino acid sequences and demonstrated that the A. peninsulae genome from BI was more closely grouped with two Korean samples (HQ660074 and JN546584) than the Chinese (KP671850) sample. This study verified the evolutionary status of A. peninsulae inhabiting the BI at the molecular level, and could be a significant supplement to the genetic background.
Abstract
The Korean field mouse, Apodemus peninsulae mitochondrial genome has previously been reported for mice obtained from mainland Korea and China. In this investigation the complete mitochondrial genome sequence for a mouse obtained from Baengnyeong Island (BI) in South Korea was determined using high-throughput whole-genome sequencing for the first time. The circular genome was determined to be 16,268 bp in length. It was found to be composed of a typical complement gene that encodes 13 protein subunits of enzymes involved in oxidative phosphorylation, two ribosomal RNAs, 22 transfer RNAs, and one control region. Phylogenetic analysis involved 13 amino acid sequences and demonstrated that the A. peninsulae genome from BI was more closely grouped with two Korean samples (HQ660074 and JN546584) than the Chinese (KP671850) sample. This study verified the evolutionary status of A. peninsulae inhabiting the BI at the molecular level, and could be a significant supplement to the genetic background.


초록
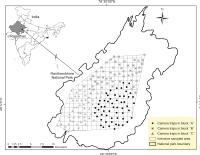
The leopard (Panthera pardus) is one of the most widespread felids worldwide. Despite their wide distribution, reliable data on leopard population densities are still inadequate for conservation and management strategies in different landscapes. In the present study, we estimated leopard density using camera traps in the Ranthambhore Tiger Reserve (RTR), Rajasthan, India, between December 2010 and February 2011, where leopards coexist alongside a high density of tigers (Panthera tigris), a larger predator (RTR). A sampling effort of 4,450 trap days was made from 178 camera trapping stations over 75 days, resulting in 46 suitable photo captures (25 right flanks and 21 left flanks). In total, 18 individuals (7 males, 8 females, and 3 unknown sexes) were identified using the right flanks, and the estimated leopard density was 8.8 (standard error=2.8) individuals/100 km2. Leopard density appeared to respond to small prey (<50 kg weight) richness. As this is the first systematic study to provide baseline information on leopard density in RTR, it could form a baseline for comparison in future investigations.
Abstract
The leopard (Panthera pardus) is one of the most widespread felids worldwide. Despite their wide distribution, reliable data on leopard population densities are still inadequate for conservation and management strategies in different landscapes. In the present study, we estimated leopard density using camera traps in the Ranthambhore Tiger Reserve (RTR), Rajasthan, India, between December 2010 and February 2011, where leopards coexist alongside a high density of tigers (Panthera tigris), a larger predator (RTR). A sampling effort of 4,450 trap days was made from 178 camera trapping stations over 75 days, resulting in 46 suitable photo captures (25 right flanks and 21 left flanks). In total, 18 individuals (7 males, 8 females, and 3 unknown sexes) were identified using the right flanks, and the estimated leopard density was 8.8 (standard error=2.8) individuals/100 km2. Leopard density appeared to respond to small prey (<50 kg weight) richness. As this is the first systematic study to provide baseline information on leopard density in RTR, it could form a baseline for comparison in future investigations.



초록
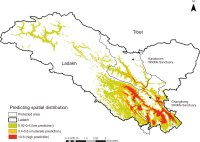
The Tibetan Plateau is home to the only alpine crane species, the black-necked crane (Grus nigricollis). Conservation efforts are severely hampered by a lack of knowledge on the spatial distribution and breeding habitats of this species. The ecological niche modeling framework used to predict the spatial distribution of this species, based on the maximum entropy and occurrence record data, allowed us to generate a species-specific spatial distribution map in Ladakh, Trans-Himalaya, India. The model was created by assimilating species occurrence data from 486 geographical sites with 24 topographic and bioclimatic variables. Fourteen variables helped forecast the distribution of black-necked cranes by 96.2%. The area under the curve score for the model training data was high (0.98), indicating the accuracy and predictive performance of the model. Of the total study area, the areas with high and moderate habitat suitability for black-necked cranes were anticipated to be 8,156 km2 and 6,759 km2, respectively. The area with high habitat suitability within the protected areas was 5,335 km2. The spatial distribution predicted using our model showed that the majority of speculated conservation areas bordered the existing protected areas of the Changthang Wildlife Sanctuary. Hence, we believe, that by increasing the current study area, we can account for these gaps in conservation areas, more effectively.
Abstract
The Tibetan Plateau is home to the only alpine crane species, the black-necked crane (Grus nigricollis). Conservation efforts are severely hampered by a lack of knowledge on the spatial distribution and breeding habitats of this species. The ecological niche modeling framework used to predict the spatial distribution of this species, based on the maximum entropy and occurrence record data, allowed us to generate a species-specific spatial distribution map in Ladakh, Trans-Himalaya, India. The model was created by assimilating species occurrence data from 486 geographical sites with 24 topographic and bioclimatic variables. Fourteen variables helped forecast the distribution of black-necked cranes by 96.2%. The area under the curve score for the model training data was high (0.98), indicating the accuracy and predictive performance of the model. Of the total study area, the areas with high and moderate habitat suitability for black-necked cranes were anticipated to be 8,156 km2 and 6,759 km2, respectively. The area with high habitat suitability within the protected areas was 5,335 km2. The spatial distribution predicted using our model showed that the majority of speculated conservation areas bordered the existing protected areas of the Changthang Wildlife Sanctuary. Hence, we believe, that by increasing the current study area, we can account for these gaps in conservation areas, more effectively.



초록
Climate change is more rapid in the Arctic than elsewhere in the world, and increased precipitation and warming are expected cause changes in biogeochemical processes due to altered microbial communities and activities. It is crucial to investigate microbial responses to climate change to understand changes in carbon and nitrogen dynamics. We investigated the effects of increased temperature and precipitation on microbial biomass and community structure in dry tundra using two depths of soil samples (organic and mineral layers) under four treatments (control, warming, increased precipitation, and warming with increased precipitation) during the growing season (June–September) in Cambridge Bay, Canada (69°N, 105°W). A phospholipid fatty acid (PLFA) analysis method was applied to detect active microorganisms and distinguish major functional groups (e.g., fungi and bacteria) with different roles in organic matter decomposition. The soil layers featured different biomass and community structure; ratios of fungal/bacterial and gram-positive/-negative bacteria were higher in the mineral layer, possibly connected to low substrate quality. Increased temperature and precipitation had no effect in either layer, possibly due to the relatively short treatment period (seven years) or the ecosystem type. Mostly, sampling times did not affect PLFAs in the organic layer, but June mineral soil samples showed higher contents of total PLFAs and PLFA biomarkers for bacteria and fungi than those in other months. Despite the lack of response found in this investigation, long-term monitoring of these communities should be maintained because of the slow response times of vegetation and other parameters in high-Arctic ecosystems.
Abstract
Climate change is more rapid in the Arctic than elsewhere in the world, and increased precipitation and warming are expected cause changes in biogeochemical processes due to altered microbial communities and activities. It is crucial to investigate microbial responses to climate change to understand changes in carbon and nitrogen dynamics. We investigated the effects of increased temperature and precipitation on microbial biomass and community structure in dry tundra using two depths of soil samples (organic and mineral layers) under four treatments (control, warming, increased precipitation, and warming with increased precipitation) during the growing season (June–September) in Cambridge Bay, Canada (69°N, 105°W). A phospholipid fatty acid (PLFA) analysis method was applied to detect active microorganisms and distinguish major functional groups (e.g., fungi and bacteria) with different roles in organic matter decomposition. The soil layers featured different biomass and community structure; ratios of fungal/bacterial and gram-positive/-negative bacteria were higher in the mineral layer, possibly connected to low substrate quality. Increased temperature and precipitation had no effect in either layer, possibly due to the relatively short treatment period (seven years) or the ecosystem type. Mostly, sampling times did not affect PLFAs in the organic layer, but June mineral soil samples showed higher contents of total PLFAs and PLFA biomarkers for bacteria and fungi than those in other months. Despite the lack of response found in this investigation, long-term monitoring of these communities should be maintained because of the slow response times of vegetation and other parameters in high-Arctic ecosystems.

_학술지_디자인_시안_koar_top_300x55.png)
