 ISSN : 2765-2203
ISSN : 2765-2203
Vol.5 No.3
Abstract
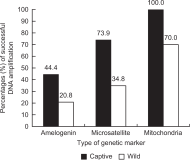
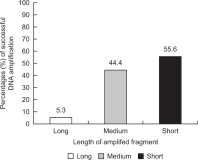
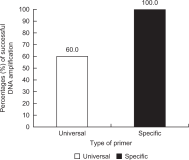
In this review, we compared the success rates of DNA amplification and introduced the efficient non-invasive sampling of fecal samples collected from captive and wild Korean long-tailed gorals (Naemorhedus caudatus) by referring to previous non-invasive studies, including three important references (Kim et al., 2008; Kim, 2021; Kim, 2022). A large difference in PCR success rates in the captive and wild populations was observed for mitochondrial (100 and 70.0%), sex-linked (44.4 and 20.8%), and microsatellite markers (73.9 and 34.8%, respectively). Out of the three types of genetic markers, the mitochondrial maker showed the highest success rate, followed by microsatellite and sex-linked markers. In addition, we estimated two factors that affected the PCR success, including the length of the amplified fragments (long, medium, and short) and the type of primer (universal and specific) in fecal samples from a captive population. The length of the PCR fragment was inversely proportional to the PCR success (5.3, 44.4, and 55.6% for long, medium, and short fragments, respectively), and the specific primer set (100%) was more efficient than the universal primer set (60.0%). This review is fundamental but would be greatly helpful for new non-invasive conservation genetic studies, particularly those that use fecal samples from captive and wild populations of rare endangered species. We recommend beginning conservation genetic experiments using mitochondrial markers and then nuclear markers, such as microsatellite and sex-linked markers, to save time, costs, and labor.


Abstract
The purpose of this study is to propose a new method of analysis focusing on interconnections between species rather than traditional biodiversity analysis, which represents ecosystems in terms of species and individual counts such as species diversity and species richness. This new approach aims to enhance our understanding of ecosystem networks. Utilizing data from the 4th National Natural Environment Survey (2014-2018), the following eight taxonomic groups were targeted for our study: herbaceous plants, woody plants, butterflies, Passeriformes birds, mammals, reptiles & amphibians, freshwater fishes, and benthonic macroinvertebrates. A co-occurrence frequency analysis was conducted using nationwide data collected over five years. As a result, in all eight taxonomic groups, the degree value represented by a linear regression trend line showed a slope of 0.8 and the weighted degree value showed an exponential nonlinear curve trend line with a coefficient of determination (R²) exceeding 0.95. The average value of the clustering coefficient was also around 0.8, reminiscent of well-known social phenomena. Creating a combination set from the species list grouped by temporal information such as survey date and spatial information such as coordinates or grids is an easy approach to discern species distributed regionally and locally. Particularly, grouping by species or taxonomic groups to produce data such as co-occurrence frequency between survey points could allow us to discover spatial similarities based on species present. This analysis could overcome limitations of species data. Since there are no restrictions on time or space, data collected over a short period in a small area and long-term national-scale data can be analyzed through appropriate grouping. The co-occurrence frequency analysis enables us to measure how many species are associated with a single species and the frequency of associations among each species, which will greatly help us understand ecosystems that seem too complex to comprehend. Such connectivity data and graphs generated by the co-occurrence frequency analysis of species are expected to provide a wealth of information and insights not only to researchers, but also to those who observe, manage, and live within ecosystems.




Abstract
Unintentional dispersal of organisms has explosively increased due to expansion of human activities. Among introduced organisms, some organisms are categorized as invasive species because of their effects on environmental risk, economic loss, and human health. In 2023, a leopard slug (Limax maximus) was reported in Suwon, Republic of Korea. This slug was designated as a potential invasive species because a wide range of plant species were identified as food sources for this slug in its original habitats. However, it is difficult to investigate the ecological risk of this newly introduced slug in Republic of Korea. Therefore, the potential ecological risk from this newly introduced slug was estimated by meta-genome analyses of its feces. Through meta-genome analyses, 22 Families, 28 Genera, and 26 Species of land plants were identified. Among these 26 identified plant species, six economically important crops – squash (Cucurbita maxima), tomato (Solanum lycopersicum), potato (Solanum tuberosum), cowpea (Vigna unguiculata), rice (Oryza sativa), and oriental melon (Cucumis melo) – were identified. Therefore, leopard slugs potentially could cause economic losses in Republic of Korea. Further study is required to build a control strategy against leopard slugs.

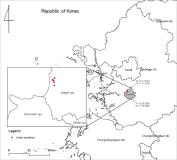


Abstract
Field surveys for reports on suspected invasive termite species were received at the National Institute of Ecology’s Invasive Species Reporting Center. We collected 10 termites and performed DNA sequence analysis for species identification. Specimens were confirmed as Coptotermes formosanus. This is the reconfirmation of C. formosanus in South Korea, highlighting the importance of early detection and rapid response and reaffirming the possibility of C. formosanus invading South Korea.



